Introduction¶
In this tutorial you will learn how to make use of the alternative starts functionality in FastROCS TK. This functionality was introduced in the OpenEye Toolkits v2017.Feb release of FastROCS TK 1.8.1.
Note
The data used in this tutorial is not provided.
Alternative starting points refer to the starting orientations of the
query-conformer
pair when optimizing their overlap in space. The default method is
OEFastROCSOrientation.Inertial. In this method, the
molecules are centered and set to an inertial frame of reference. The database
molecule is then rotated about its inertial axes, creating up to a possible 8
starting points at which overlays are then optimized.
The default optimizes the first
4 inertial starts only. This can be changed via the
OEShapeDatabaseOptions.SetNumInertialStarts method.
The figure below shows the inertial starting points of caffeine.
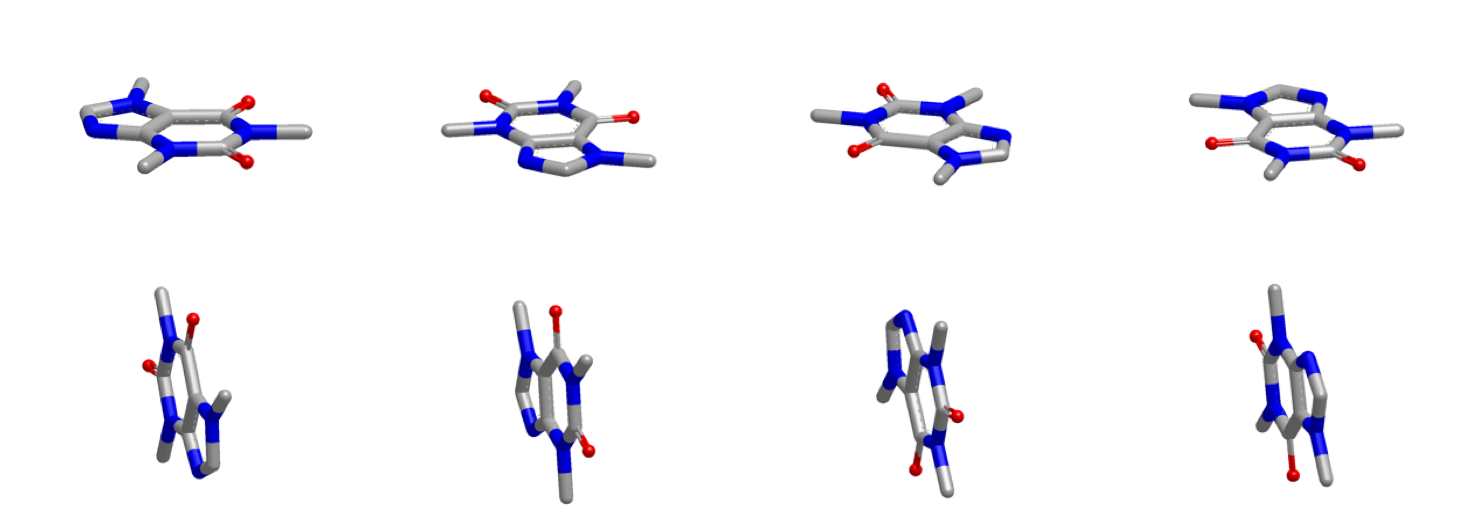
Inertial Starting Points of Caffeine¶
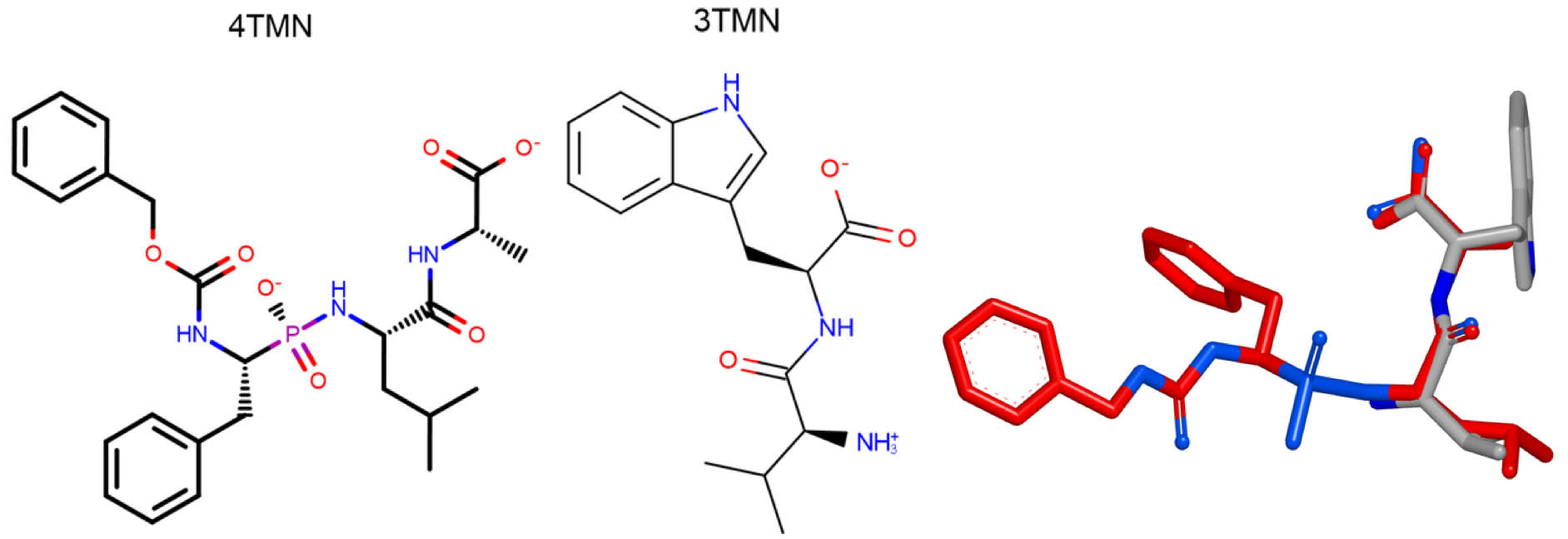
For some molecular structures these inertial starting positions may not lead to adequate sampling of the molecular volume overlap space, resulting in poor overlap/scores. Making use of alternative starts can solve this problem by creating additional starting positions at which to perform the inertial optimizations. One example of this is the alignment of 4TMN and 3TMN ligands of Thrombin [Grant-1996].
The figure below shows the 2D molecular structure of these ligands and their x-ray alignment. The x-ray alignment shows a good overlap between 3TMN and the side chain of 4TMN.
The figure below shows the resultant overlays in FastROCS TK
when using default starting points compared to
OEFastROCSOrientation.InertialAtHeavyAtoms
starting points.
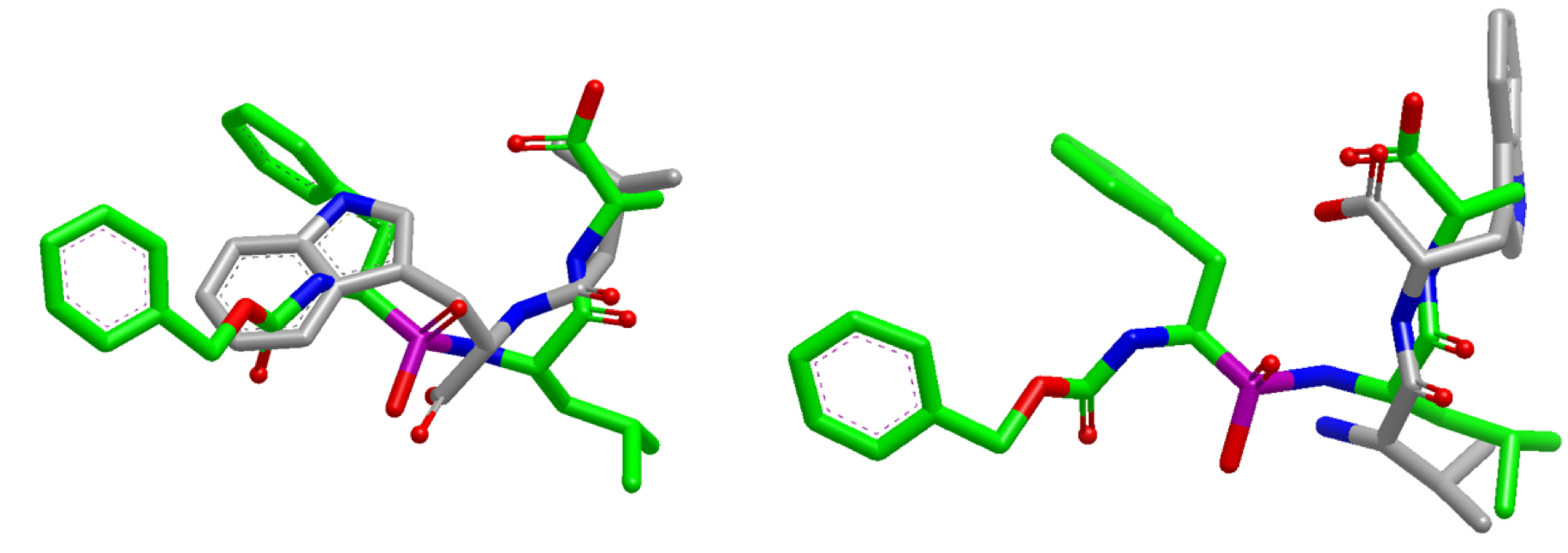
Overlap Comparison¶
Overlay generated from performing a default search (left) compared with ”InertialAtHeavyAtoms” search (right).
The default search results in poor overlap between the two molecules (left).
When the new
OEFastROCSOrientation.InertialAtHeavyAtoms
orientation option is used, the resultant overlap is much more
favorable (right) and correlates well with the x-ray alignment
from Alternative Starts Use Case.
There are 5 methods currently implemented in FastROCS TK that allow different starting orientations to be experimented with. They are:
A description of these can be found in the
OEFastROCSOrientation section of the docs.
This tutorial will focus on 3 alternative starts methods:
The methodology described can be adapted to the remaining starting point
options that are not described in this tutorial. For example, the use case for
OEFastROCSOrientation.InertialAtColorAtoms
is almost identical to
OEFastROCSOrientation.InertialAtHeavyAtoms except for
the constant passed in to
OEShapeDatabaseOptions.SetInitialOrientation
and the use of
OEShapeDatabaseOptions.GetNumColorAtomStarts instead of
OEShapeDatabaseOptions.GetNumHeavyAtomStarts
to retrieve the number of starts.