Aromaticity Perception¶
Aromaticity and Hückel’s rule¶
OEChem TK’s aromaticity perception routines are based around the Hückel’s rule that defines cyclic conjugated systems with \((4N+2)\) number of \(\pi\) electrons as aromatic (where \(N\) is zero or any positive integer).
Aromaticity can be set using the
OEAssignAromaticFlags function,
which takes an OEMolBase argument.
The OEAssignAromaticFlags sets
the aromaticity flags on atoms and bonds using an aromaticity model.
(For the list of available aromaticity models in OEChem TK* see
section Aromaticity Models in OEChem TK. )
mol = oechem.OEGraphMol()
oechem.OEParseSmiles(mol, "C1[NH]C=CC=1CO")
oechem.OEAssignAromaticFlags(mol)
Hint
The OESmilesToMol function automatically perceives
the aromaticity of the molecule using the default
OEAroModel_OpenEye aromaticity model.
The following two code snippets demonstrate how to loop over aromatic atoms
using the OEIsAromaticAtom functor and
the IsAromatic method of
the OEAtomBase class.
for atom in mol.GetAtoms(oechem.OEIsAromaticAtom()):
print(atom.GetIdx(), oechem.OEGetAtomicSymbol(atom.GetAtomicNum()))
for atom in mol.GetAtoms():
if atom.IsAromatic():
print(atom.GetIdx(), oechem.OEGetAtomicSymbol(atom.GetAtomicNum()))
The aromatic bonds of a molecule can similarly be accessed using the
OEIsAromaticBond functor and the
IsAromatic method of the
OEBondBase class.
For more information about functors see chapter Predicate Functors.
The user can also set the atom and bond aromaticity flags manually using the
OEAtomBase.SetAromatic and
OEBondBase.SetAromatic methods.
Aromaticity Models in OEChem TK¶
The OEAssignAromaticFlags function can
also take an aromaticity model constant as an argument to perceive various
aromaticity models.
The following aromaticity models are available in OEChem TK:
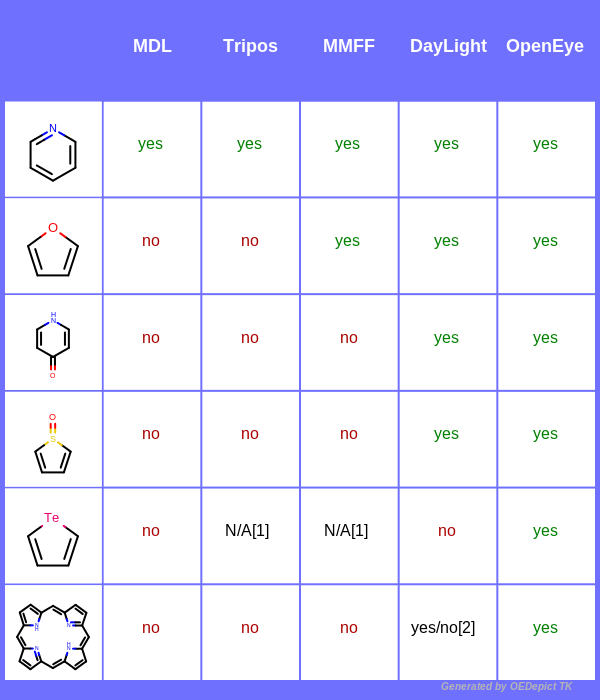
The following table demonstrates the differences between the five available aromaticity models.
Table footnotes:
[1] Atomic elements such as Te, B, Se are not available in the MMFF and Tripos aromaticity models.
[2] Only two out of the four five-membered rings are recognized as aromatic.
The code in Listing 1 demonstrates how to perceive aromaticity
with these available models.
Listing 1: Aromaticity perception with various models
from openeye import oechem
def PerceiveAromaticity(mol, modelname, aromodel):
oechem.OEAssignAromaticFlags(mol, aromodel)
cansmi = oechem.OECreateCanSmiString(mol)
print(modelname, ":", cansmi)
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "c1ncncc1c2cocc2-c3[nH]ccc(=O)c3")
PerceiveAromaticity(mol, "OEAroModelOpenEye ", oechem.OEAroModel_OpenEye)
PerceiveAromaticity(mol, "OEAroModelDaylight", oechem.OEAroModel_Daylight)
PerceiveAromaticity(mol, "OEAroModelTripos ", oechem.OEAroModel_Tripos)
PerceiveAromaticity(mol, "OEAroModelMMFF ", oechem.OEAroModel_MMFF)
PerceiveAromaticity(mol, "OEAroModelMDL ", oechem.OEAroModel_MDL)
Since these models define aromaticity rules differently, the generated canonical SMILES
depend on the applied aromaticity models.
The output of Listing 1 is the following:
OEAroModel::OpenEye : c1c[nH]c(cc1=O)c2cocc2c3cncnc3
OEAroModel::Daylight : c1c[nH]c(cc1=O)c2cocc2c3cncnc3
OEAroModel::Tripos : c1c(cncn1)C2=COC=C2C3=CC(=O)C=CN3
OEAroModel::MMFF : c1c(cncn1)c2cocc2C3=CC(=O)C=CN3
OEAroModel::MDL : c1c(cncn1)C2=COC=C2C3=CC(=O)C=CN3
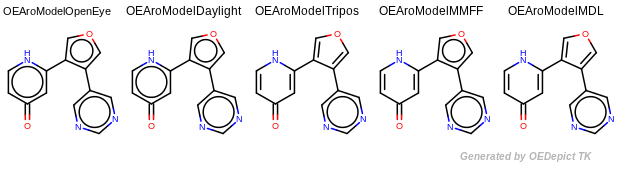
Example of aromaticity perception with different models¶
Clearing Aromaticity¶
The aromatic property of all atoms and bonds in a molecule, can conveniently be
cleared (i.e. set to value false) by calling the
OEClearAromaticFlags function.
This is useful when writing the Kekulé form of a SMILES string, which can be done
by calling OEClearAromaticFlags before
calling OEKekulize and
OECreateSmiString functions.
Listing 2: Clearing aromaticity
from openeye import oechem
mol = oechem.OEGraphMol()
oechem.OEParseSmiles(mol, "n1ccncc1")
print("Canonical smiles :", oechem.OECreateCanSmiString(mol))
oechem.OEClearAromaticFlags(mol)
oechem.OEKekulize(mol)
print("Kekule smiles :", oechem.OECreateCanSmiString(mol))
The output of Listing 2 is the following:
Canonical smiles : c1cnccn1
Kekule smiles : C1=CN=CC=N1
Input/Output Aromaticity¶
Since OEParseSmiles preserves the aromaticity
present (or absent) in the input SMILES string,
the OEClearAromaticFlags or
the OEAssignAromaticFlags have to be explicitly
called to remove or perceive aromaticity in a molecule, respectively.
(See examples in Listing 1 and Listing 2)
However, when a molecule is imported from a file with a high-level
OEReadMolecule function,
atom and bond aromaticity is automatically perceived using the default
OEAroModelOpenEye model.
As mentioned before (in section Molecular Property Preservation), the
high-level OEWriteMolecule` writer function
may automatically update atom and bond properties (including aromaticity)
in order to standardize the exported molecules.
The following table shows the aromaticity models associated with various file
formats.
File Format |
Default Output Aromaticity Models |
|---|---|
[1] |
|
[1] |
|
[1] |
|
[1] |
|
[1] |
|
[1] |
|
[1] |
|
[1] |
|
[1] |
Table footnote:
[1] The aromaticity model is not changed by the associated molecule writer.
The following snippet shows how to overwrite the default aromaticity model of a specific file format.
ofs = oechem.oemolostream(".smi")
oechem.OEWriteMolecule(ofs, mol) # using default OpenEye aromaticity model
flavor = ofs.GetFlavor(ofs.GetFormat())
flavor |= oechem.OEOFlavor_Generic_OEAroModelMDL
ofs.SetFlavor(ofs.GetFormat(), flavor)
oechem.OEWriteMolecule(ofs, mol)
This will produce the following output:
c1c[nH]cc1CO
C1=CNC=C1CO
See also