Connectivity Perception¶
OEChem TK provides several functions for determining the connectivity and/or bond orders from various input file formats. For correct molecule processing, OEChem TK requires all the covalent bonds to be represented in a molecule and each bond to have a defined bond order, 1 for single, 2 for double, 3 for triple and 4 for quadruple. Given this explicit Kekulé representation of a molecule, OEChem TK can perceive and re-perceive high order attributes such as ring membership or aromaticity as defined by different aromaticity models.
Alas, unlike MDL’s SD file format, not all file formats explicitly specify a Kekulé form of a molecule with explicit bond orders. The functions, described in this chapter, attempt to deduce such a representation from the information that is available in such file formats.
Connectivity From 3D Coordinates¶
For file formats that provide 3D coordinates, but not explicit bond information
(or only partial bond information), OEChem TK uses the
OEDetermineConnectivity function.
This function deduces the pattern of covalent bonding in a molecule from the
proximity of atoms.
Two atoms are considered bonded if they are located within the sum of their
covalent radii (OEGetCovalentRadius)
plus an additional “slop” factor of 0.45 Angstroms.
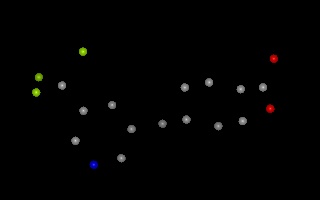
Example of 3D molecule with no explicit bond information¶
OEDetermineConnectivity will not
create a bond between two atoms that are less than 0.4 Angstroms apart.
Such unreasonably short bond lengths indicate the structure is either severely
distorted, or doesn’t have coordinate information at all.
All bonds created by OEDetermineConnectivity
have bond orders set to one.
To perceive bond order information, see OEChem TK’s
OEPerceiveBondOrders function described
in the next section.
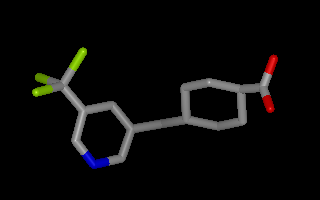
Example of 3D molecule with perceived bond connectivity¶
The OEDetermineConnectivity function
checks whether a bond already exists between two atoms before creating a new
bond.
This allows this function to be used with file formats that may specify partial
connectivity, such as only multiple (double, triple or quadruple) bonds.
Bond Order Perception¶
The OEPerceiveBondOrders function is
used to deduce bond orders from the 3D coordinates and simple connectivity
of a molecule.
If the simple connectivity, i.e. bonds without bond orders isn’t specified in
the input file, OEDetermineConnectivity
should be called first to deduce this information from the 3D coordinates.
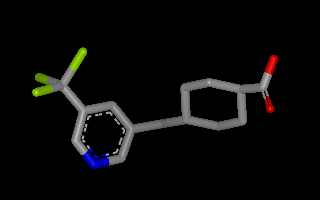
Example of 3D molecule with perceived connectivity and bond order¶
The following code snippet shows how to perceive connectivity and bond order if a molecule has 3D information but no explicit bond information:
oechem.OEDetermineConnectivity(mol)
oechem.OEFindRingAtomsAndBonds(mol)
oechem.OEPerceiveBondOrders(mol)
oechem.OEAssignImplicitHydrogens(mol)
oechem.OEAssignFormalCharges(mol)
Kekulé Form Assignment¶
A number of file formats don’t represent a connection table as a
single representative Kekulé form but instead denote some bonds,
such as those in benzene, as aromatic. OEChem TK provides a method for
determining a valid, but arbitrary, Kekulé form for such connection
tables using the OEKekulize function
(see example in Figure: Kekulization of
quinolin). On input to
OEKekulize, the integer bond type
property of each bond represents either the bond order (1 for single,
2 for double, 3 for triple or 4 for quadruple) or the value 5
indicating the bond is aromatic or resonant. The algorithm sets the
bond order property from the bond type property, with the exception of
bond type 5, which is assigned a bond order of either 1 or 2
representing either a single or double bond. The boolean return value
indicates whether a valid Kekulé form could be assigned.
See also
OEKekulize function is normally only used by
low-level file readers for interpreting input connection tables.
To write out a Kekulé SMILES string, the aromaticity atom and bond flags
have to be cleared by the OEClearAromaticFlags
function in order to consider a molecule as aliphatic with explicit bond order.
See example code in Clearing Aromaticity.
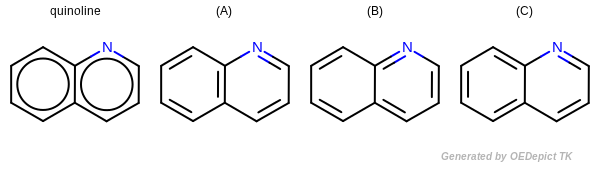
Kekulization of quinolin¶
OEKekulize() function arbitrary generates one of the three valid Kekulé forms
Kekulé Assignment of New Bonds¶
Molecules input to
OEKekulize must have their integer bond type set.
The property of each bond represents either the bond order (1 for single,
2 for double, 3 for triple or 4 for quadruple) or the value 5 indicating
the bond is aromatic or resonant. When creating new atoms and bonds internally
the integer type is set to a default value of 0. The following code snippet
outlines how to prepare modified molecules by setting the bond int type so that
they can be passed to OEKekulize:
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "c1ccccc1")
# Code that modifies the molecule
# and creates new atoms and bonds
oechem.OEFindRingAtomsAndBonds(mol)
oechem.OEAssignAromaticFlags(mol)
for bond in mol.GetBonds():
if bond.IsAromatic():
bond.SetIntType(5)
elif bond.GetOrder() != 0:
bond.SetIntType(bond.GetOrder())
else:
bond.SetIntType(1)
oechem.OEKekulize(mol)