FastROCS UI¶
FastROCS has a simple interface. To search, we need a query molecule. The query is normally a 3D structure if known, but a 2D structure will be automatically converted to 3D before the search.
To get started, click the “New Query” button.
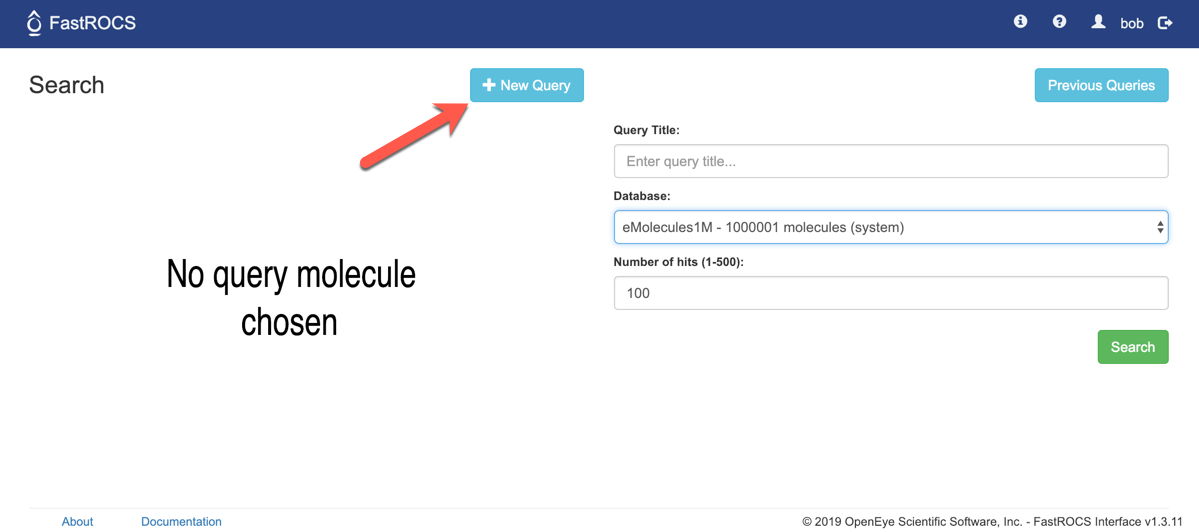
There are 3 ways to enter queries.
First, we have a full-featured sketcher allowing creation of queries from scratch. The sketcher also supports paste as long as you copy the molecule as a MOL file or SMILES. For BioviaDraw, use Ctrl+M, for example.
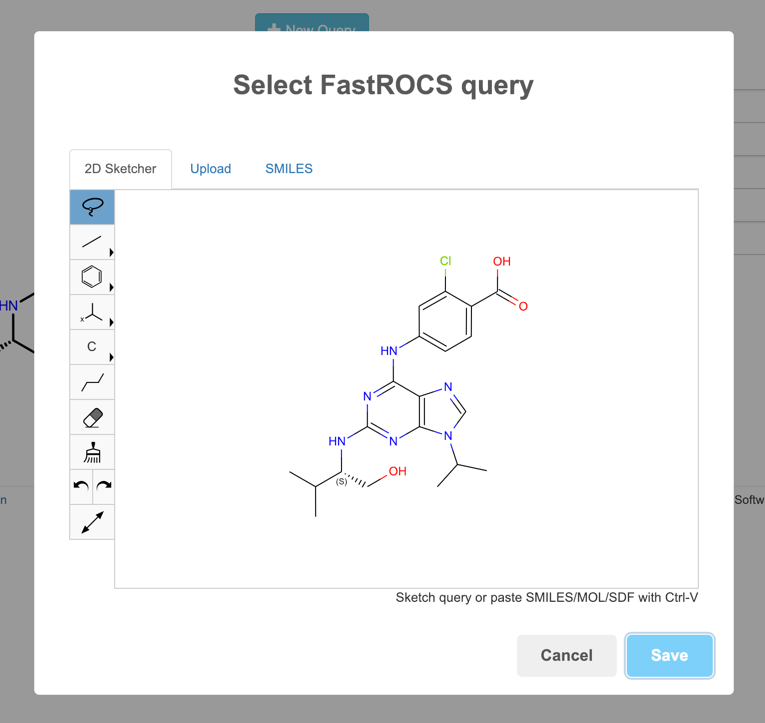
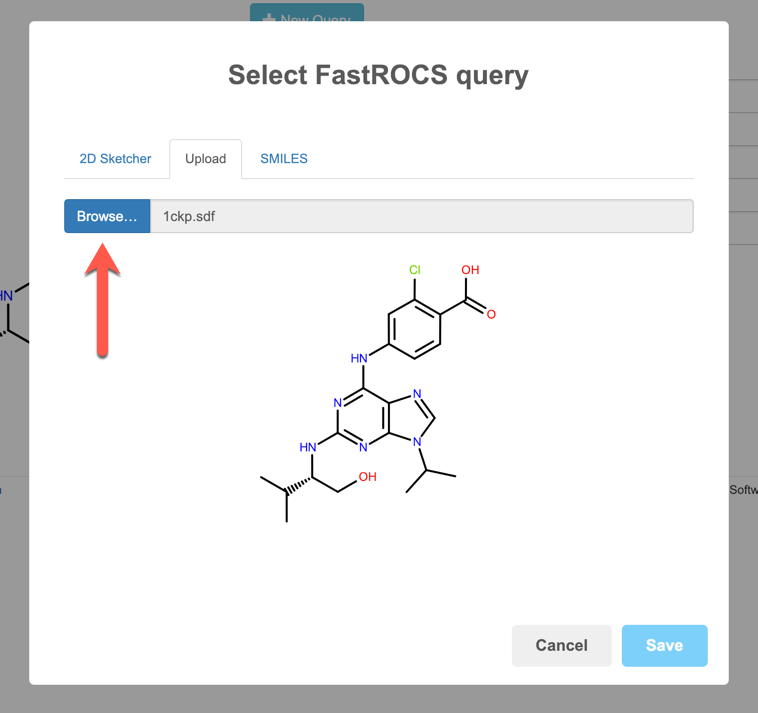
For a 3D query, use a local molecule file. Click the Browse button, navigate to the file of interest and hit Save.
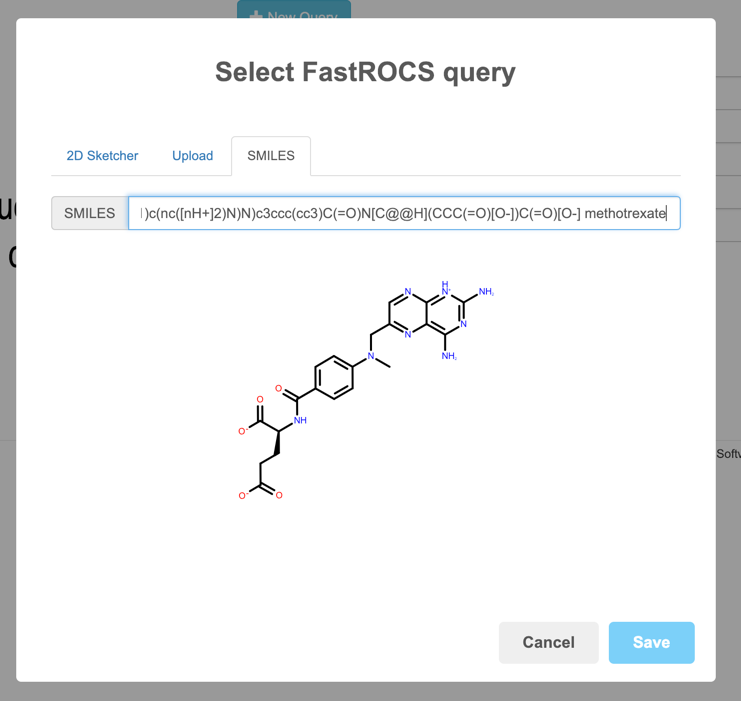
SMILES can be entered directly as well.
Once you have a query, choose one of the available databases to search and then hit the Search button.
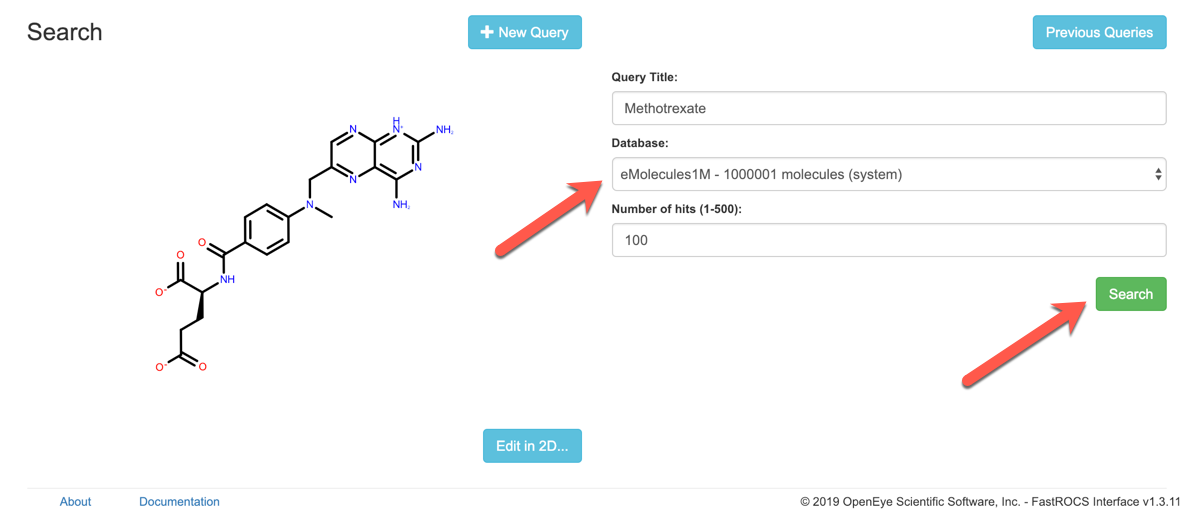
While the search is queued or running, you will see a status page. Once the search is complete, the browser will be re-directed to the results page.
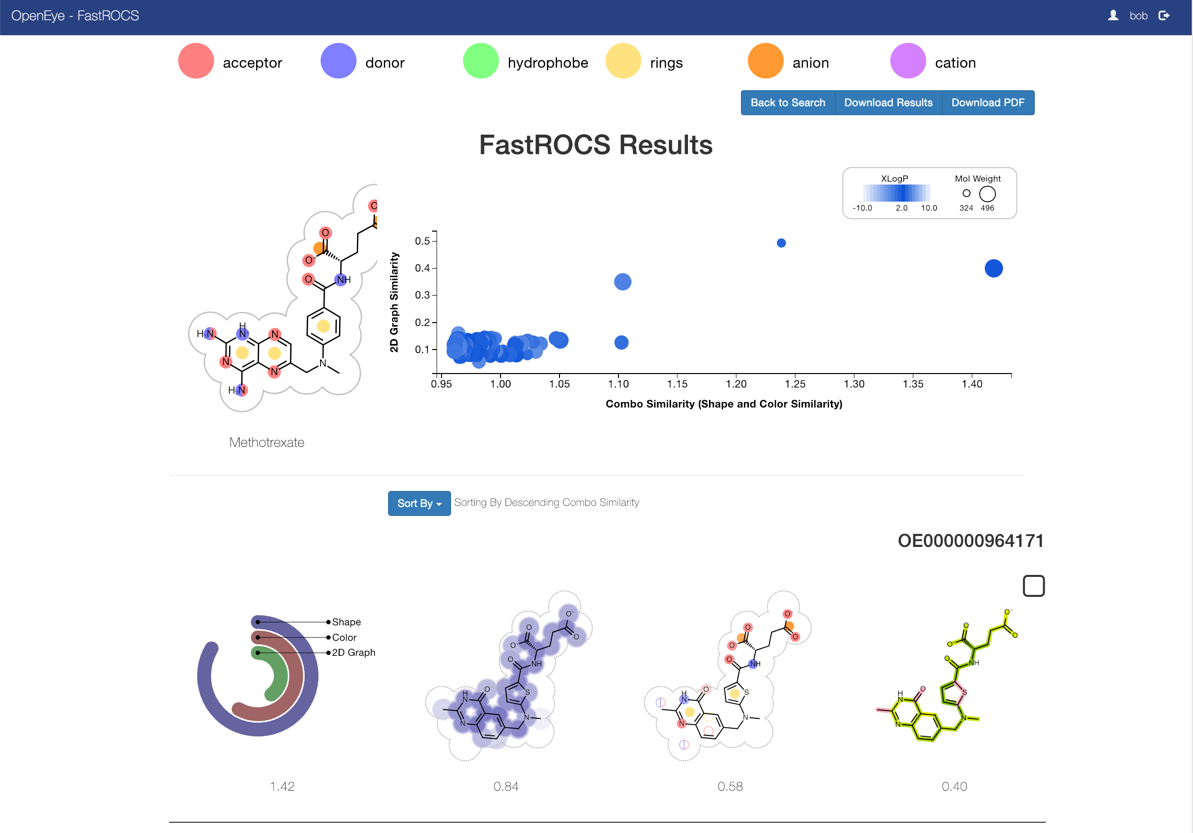
On the results page, you can view 2D depictions of the shape and color overlaps along with a measure of 2D similarity.
In the upper right, there are 3 choices.
Back to Search - perform a new search, starting with this same query.
Download Results - download a molecule file of these results.
Download PDF - download a PDF representation of the 2D overlap depictions.
Back on the Search page, we have 2 other options.
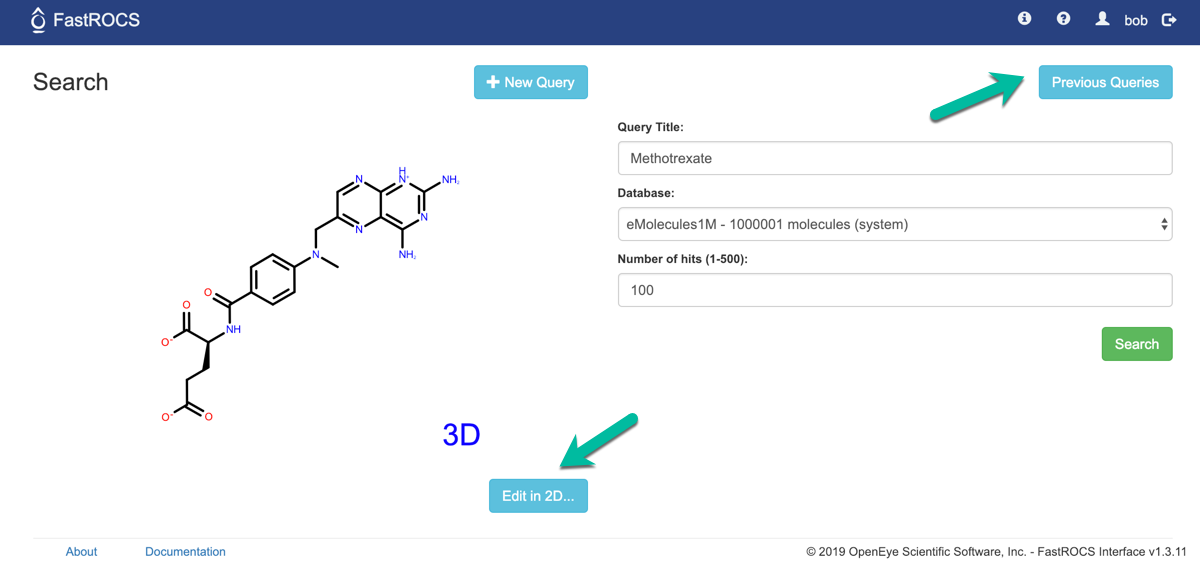
Edit in 2D … - open the sketcher with the current query loaded
Previous Queries - go to page showing all previous queries
Previous queries¶
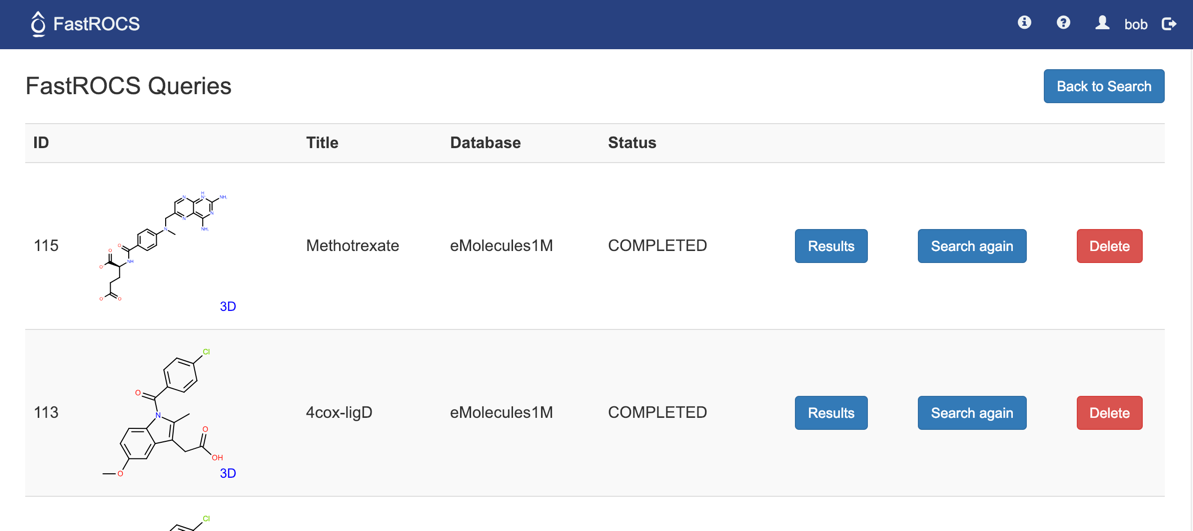
For each query:
Results - Load the results page for this query.
Search Again - Return to Search page and load this query.
Delete - Delete this query and its results from server.