MMDS Data Preparation¶
MMDS loads experimental protein structure files, along with their X-ray data or cryo-EM data. While there is a client to load structures in pieces, setting up the initial data set for bulk preparation and upload will speed up the first time.
A worked example¶
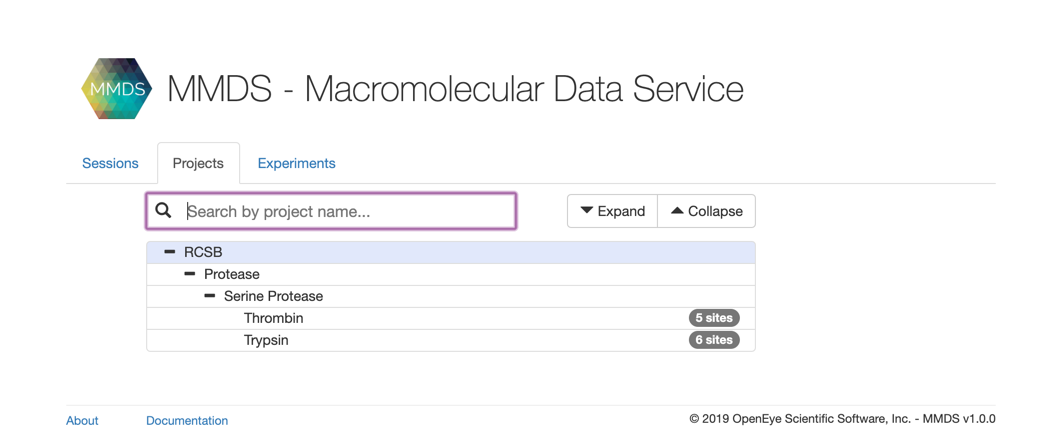
We will use a couple if example families to create a set of structures for MMDS. Working through this example should highlight the basic process that can be extended to more families of interest.
We are going to use two Serine Protease families, Trypsin and Thrombin.
Data Preparation¶
The first thing to do is to gather the structures, density files and other information for these two projects. We will then use Spruce to prepare OEDesignUnits and then finally load these results into MMDS.
$ mkdir mmds-data
$ cd mmds-data
Putting some PDB codes into a text file, for Trypsin and Thrombin, we can use Spruce to download the structures and density files.
1G3D
1GJ6
1K1L
1GHZ
1TRN
3RML
3DA9
3P17
3QWC
4BAH
Use spruce getpdb to get the structure files into a directory for each list, then use spruce getmap to get the density files for each structure file.
$ spruce getpdb --list trypsin_pdb_codes.list -o trypsin -v
1G3D: SUCCESS
1GJ6: SUCCESS
1K1L: SUCCESS
1GHZ: SUCCESS
1TRN: SUCCESS
$ spruce getmap --dir trypsin -v
1g3d: wrote trypsin/1g3d.mtz
1ghz: wrote trypsin/1ghz.mtz
1gj6: wrote trypsin/1gj6.mtz
1k1l: wrote trypsin/1k1l.mtz
1trn: wrote trypsin/1trn.mtz
$ spruce getpdb --list thrombin_pdb_codes.list -o thrombin -v
3RML: SUCCESS
3DA9: SUCCESS
3P17: SUCCESS
3QWC: SUCCESS
4BAH: SUCCESS
$ spruce getmap --dir thrombin -v
3da9: wrote thrombin/3da9.mtz
3p17: wrote thrombin/3p17.mtz
4bah: wrote thrombin/4bah.mtz
3qwc: wrote thrombin/3qwc.mtz
3rml: wrote thrombin/3rml.mtz
Meta data¶
For each structure, we want to create an extra file containing meta data
associated with it. This includes info on the the author, date, method and
ligand HET info. For public data, we can create these files from the PDB
header info. For private/internal structure, use the
OEStructureMetaData class.
An example meta data file:
{
"Author": "E.TOYOTA,K.K.S.NG,H.SEKIZAKI,K.ITOH,K.TANIZAWA,M.N.G.JAMES",
"Keywords": [
"ENZYME-INHIBITOR COMPLEX",
" COORDINATION METAL BASED INHIBITOR",
"HYDROLASE"
],
"ExperimentType": "X-RAY DIFFRACTION",
"ExperimentDate": "17-JAN-01",
"Revision": "1.3",
"RevisionDate": "04-APR-18",
"IridiumData": {
"Category": 0,
"LaD": 0,
"ASaD": 0,
"POL": false,
"POAS": false,
"AltConfs": false,
"PackRes": false,
"Excp": false,
"IrrRFree": false,
"PossCov": false,
"DPI": 0.0897862915057751,
"RFree": 0.189,
"Resolution": 1.8,
"HasMTZ": false
},
"SequenceMetadata": [
{
"ChainID": "A",
"StartResNum": 11,
"StartResInsCode": " ",
"Sequence": "ASP-ASP-ASP-ASP-LYS-ILE-VAL-GLY-GLY-TYR-THR-CYS-GLY-ALA-ASN-THR-VAL-PRO-TYR-GLN-VAL-SER-LEU-ASN-SER-GLY-TYR-HIS-PHE-CYS-GLY-GLY-SER-LEU-ILE-ASN-SER-GLN-TRP-VAL-VAL-SER-ALA-ALA-HIS-CYS-TYR-LYS-SER-GLY-ILE-GLN-VAL-ARG-LEU-GLY-GLU-ASP-ASN-ILE-ASN-VAL-VAL-GLU-GLY-ASN-GLU-GLN-PHE-ILE-SER-ALA-SER-LYS-SER-ILE-VAL-HIS-PRO-SER-TYR-ASN-SER-ASN-THR-LEU-ASN-ASN-ASP-ILE-MET-LEU-ILE-LYS-LEU-LYS-SER-ALA-ALA-SER-LEU-ASN-SER-ARG-VAL-ALA-SER-ILE-SER-LEU-PRO-THR-SER-CYS-ALA-SER-ALA-GLY-THR-GLN-CYS-LEU-ILE-SER-GLY-TRP-GLY-ASN-THR-LYS-SER-SER-GLY-THR-SER-TYR-PRO-ASP-VAL-LEU-LYS-CYS-LEU-LYS-ALA-PRO-ILE-LEU-SER-ASP-SER-SER-CYS-LYS-SER-ALA-TYR-PRO-GLY-GLN-ILE-THR-SER-ASN-MET-PHE-CYS-ALA-GLY-TYR-LEU-GLU-GLY-GLY-LYS-ASP-SER-CYS-GLN-GLY-ASP-SER-GLY-GLY-PRO-VAL-VAL-CYS-SER-GLY-LYS-LEU-GLN-GLY-ILE-VAL-SER-TRP-GLY-SER-GLY-CYS-ALA-GLN-LYS-ASN-LYS-PRO-GLY-VAL-TYR-THR-LYS-VAL-CYS-ASN-TYR-VAL-SER-TRP-ILE-LYS-GLN-THR-ILE-ALA-SER-ASN"
}
],
"HeterogenMetadata": [
{
"Title": "108",
"Id": "108",
"Smiles": "C[C@@H](C(=O)O)NCc1cc(ccc1O)C(=N)N",
"Type": "unknown",
"Tautomers": []
}
]
}
Here is a small script that will create these files for all the PDB structures in one directory.
$ python create_meta_data.py --dir trypsin -v
created trypsin/1K1L.pdb.json
created trypsin/1G3D.pdb.json
created trypsin/1GHZ.pdb.json
created trypsin/1GJ6.pdb.json
created trypsin/1TRN.pdb.json
$ python create_meta_data.py --dir thrombin -v
created thrombin/3DA9.pdb.json
created thrombin/3P17.pdb.json
created thrombin/3QWC.pdb.json
created thrombin/4BAH.pdb.json
created thrombin/3RML.pdb.json
Preparing biounits and OEDesignUnits¶
For each project, we want to use a reference OEDesignUnit to prepare all other structures. This ensures we have a consistent definition of the Bio Unit for all members and allows creating OEDesignUnits for apo structures, transferring the site info from the reference to the apo OEDesignUnit.
For Trypsin, we will use 1G3D and for Thrombin, 3RML. We will prep each of these separately and then prepare the rest using the results as reference.
$ cd trypsin
$ spruce prep -v --build-sc --cap-termini 1G3D.pdb
spruce prep run options
-- snip --
processing 1 pdb file(s) with 1 CPUs
PDB file: 1G3D.pdb
metadata file: ./1G3D.pdb.json
MTZ file: ./1g3d.mtz
Splitting ASU -> DUs
-- snip --
DU(s):
1G3D(A)__DU__biounit
1G3D(A) > 108(A-601)
Output:
wrote ./trypsin/1G3D_A__DU__biounit.oedu
wrote ./trypsin/1G3D_A__DU__108_A-601.oedu
elapsed: 8.1 secs
Creating design units: 8.17 secs
$ cd ..
Note that Spruce created 2 OEDesignUnit files. We want the one with the ligand 108 in the title, which is the known ligand in this structure.
Do the same for Thrombin/3RML:
$ cd thrombin
$ spruce prep -v --build-sc --cap-termini 3RML.pdb
-- snip --
DU(s):
3RML(HIL)__DU__biounit
3RML(HIL) > M31(H-1)
Output:
wrote ./thrombin/3RML_HIL__DU__biounit.oedu
wrote ./thrombin/3RML_HIL__DU__M31_H-1.oedu
elapsed: 20.4 secs
Creating design units: 20.42 secs
$ cd ..
Now we can prep all the other structures, using references. You may seem some warnings about charge addition or removal of clashing waters due to the addition of side chains. These are ok to ignore.
$ spruce prep --build-sc --cap-termini --design-ref trypsin/1G3D_A__DU__108_A-601.oedu --dir trypsin -o trypsin
Creating design units [*************************************************] 100.0%
Elapsed: 25.08 secs
$ ls trypsin
1G3D.pdb 1GHZ_A__DU__120_A-246.oedu 1GJ6_spruce_prep.log 1TRN.pdb.json 1ghz.mtz
1G3D.pdb.json 1GHZ_A__DU__biounit.oedu 1K1L.pdb 1TRN_A__DU__apo_A-226.oedu 1gj6.mtz
1G3D_A__DU__108_A-601.oedu 1GHZ_spruce_prep.log 1K1L.pdb.json 1TRN_A__DU__biounit.oedu 1k1l.mtz
1G3D_A__DU__biounit.oedu 1GJ6.pdb 1K1L_A__DU__FD3_A-999.oedu 1TRN_B__DU__apo_B-226.oedu 1trn.mtz
1G3D_spruce_prep.log 1GJ6.pdb.json 1K1L_A__DU__biounit.oedu 1TRN_B__DU__biounit.oedu spruce.log
1GHZ.pdb 1GJ6_A__DU__132_A-246.oedu 1K1L_spruce_prep.log 1TRN_spruce_prep.log
1GHZ.pdb.json 1GJ6_A__DU__biounit.oedu 1TRN.pdb 1g3d.mtz
$ spruce prep --build-sc --cap-termini --design-ref thrombin/3RML_HIL__DU__M31_H-1.oedu --dir thrombin -o thrombin
Creating design units [*************************************************] 100.0%
Elapsed: 26.08 secs
$ ls thrombin
3DA9.pdb 3P17_HIL__DU__biounit.oedu 3RML.pdb.json 4BAH.pdb
3DA9.pdb.json 3P17_spruce_prep.log 3RML_HIL__DU__M31_H-1.oedu 4BAH.pdb.json
3DA9_ABD__DU__44U_B-1.oedu 3QWC.pdb 3RML_HIL__DU__biounit.oedu 4BAH_ABD__DU__MEL_B-1291.oedu
3DA9_ABD__DU__biounit.oedu 3QWC.pdb.json 3RML_spruce_prep.log 4BAH_ABD__DU__biounit.oedu
3DA9_spruce_prep.log 3QWC_HIL__DU__98P_H-2001.oedu 3da9.mtz 4BAH_spruce_prep.log
3P17.pdb 3QWC_HIL__DU__biounit.oedu 3p17.mtz 4bah.mtz
3P17.pdb.json 3QWC_spruce_prep.log 3qwc.mtz spruce.log
3P17_HIL__DU__99P_H-1001.oedu 3RML.pdb 3rml.mtz
For each experiment, we now have a structure file (*.pdb), a density file (*.mtz), a meta data file (*.pdb.json), a bio unit file and at least one OEDesignUnit with a designated binding site. Time to load these into MMDS.
Experiments¶
Experiments are the core of MMDS. Generally, we are talking about X-ray crystallography experiments with their corresponding density (MTZ) map.
MMDS can also load cryo-EM (with maps), NMR and modeled structures.
Each experiment needs at least 2 pieces of data, the original structure file (PDB), and a metadata file in JSON format. If there is a map file, that is included as well. We will also upload prepared biounit files, and subsequently any sites (OEDesignUnits) created by Spruce.
The meta data file is used to include important experiment info found in the header of public structures but often stored elsewhere for proprietary structures. An important piece of this data is to provide information on HET structures in the experiment file. This includes a mapping of the 3-letter HET code in the file to an internal compound ID as well as the SMILES for each structure to help in validation.
For uploading an Experiment to MMDS, we only want the former, but later we can upload the other DU as a Site.
Note
All commands below assume an MMDS profile called ‘local’ has been created to point to your MMDS server.
Adding an experiment¶
$ mmdscli --profile local experiment list
No resources found
$ mmdscli --profile local experiment add -h
Usage: mmdscli experiment add [OPTIONS] CODE STRUCTURE META
Add a new experiment to MMDS
Options:
--density density Density/map file for this experiment
--biounit BU Biounit file. Add this flag multiple times to add all
biounits. If not provided,
mmdscli will look for spruce
created BUs in the same directory as STRUCTURE
-y, --yes Say yes to any yes/no questions. [default: False]
--json Return JSON instead of simple text on stdout.
-q, --quiet Minimal output
-v, --verbose Verbose output
-h, --help Show this message and exit.
$ cd trypsin
$ mmdscli experiment add 1G3D 1G3D.pdb 1G3D.pdb.json --density 1g3d.mtz --json
{
"DPI": 0.0897862915057751,
"RFree": 0.189,
"RFree_found": false,
"author": "E.TOYOTA,K.K.S.NG,H.SEKIZAKI,K.ITOH,K.TANIZAWA,M.N.G.JAMES",
"biounit": [
{
"experiment": 1787,
"filename": "1G3D_A__DU__biounit.oedu",
"id": 2126,
"url": "http://localhost:8080/api/v1/biounit/2126/1G3D_A__DU__biounit.oedu"
}
],
"code": "1G3D",
"created": "2019-04-16T08:57:05.539899Z",
"density": {
"filename": "1g3d.mtz",
"json": null,
"pk": 1306,
"url": "http://localhost:8080/api/v1/experiment/1787/density/1g3d.mtz"
},
"id": 1787,
"keywords": "ENZYME-INHIBITOR COMPLEX, COORDINATION METAL BASED INHIBITOR,HYDROLASE EC:3.4.21.4",
"ligands": {
"108": {
"id": "108",
"smiles": "C[C@@H](C(=O)O)NCc1cc(ccc1O)C(=N)N",
"type": ""
}
},
"method": "X-RAY DIFFRACTION",
"public": true,
"resolution": 1.8,
"sites": [],
"solved_date": "2001-01-17T00:00:00Z",
"strdate": "17-JAN-01",
"structure": {
"filename": "1G3D.pdb",
"json": "http://localhost:8080/api/v1/experiment/1787/structure/",
"pk": 5907,
"url": "http://localhost:8080/api/v1/experiment/1787/structure/1G3D.pdb"
},
"tags": []
}
$ mmdscli --profile local status
version oe_license_expires families experiments sites contexts
--------- -------------------- ---------- ------------- ------- ----------
1.0.0 2019-09-04 0 1 0 0
We can continue through each directory loading experiments one at a time, or we can use a script that uses the mmdsclient Python API to load all the experiments in a single directory at once.
Note
Since these are simple Python scripts, the MMDS_PROFILE environment variable needs to be set in order for the mmdsclient API to load the appropriate credentials.
$ MMDS_PROFILE=local python load_all_experiments.py trypsin
Found 5 input structure files
Found 5 input density files
Found 6 input biounit files
Found 5 input experiments
✔ Gathering existing experiments
Found 1 existing experiments.
Found 4 new experiments to load
loading 4 experiments [*************************************************] 100.0%
Elapsed: 3.91 secs
$ MMDS_PROFILE=local python load_all_experiments.py thrombin
Found 5 input structure files
Found 5 input density files
Found 5 input biounit files
Found 5 input experiments
✔ Gathering existing experiments : 5
Found 5 existing experiments.
Found 5 new experiments to load
loading 5 experiments [*************************************************] 100.0%
Elapsed: 4.48 secs
We can check the server status again, to see that all 10 of our experiments are loaded.
$ mmdscli --profile local status
version oe_license_expires families experiments sites contexts
--------- -------------------- ---------- ------------- ------- ----------
1.0.0 2019-09-04 0 10 0 0
We can also get a list.
Warning
Note that for 1000+ structures, this command will take some time, so be careful.
$ mmdscli --profile local experiment list --fields id,code,DPI,method,resolution
id code method DPI resolution public
---- ------ ----------------- --------- ------------ --------
1550 1G3D X-RAY DIFFRACTION 0.0897863 1.8 True
1551 1K1L X-RAY DIFFRACTION 0.308722 2.5 True
1552 1TRN X-RAY DIFFRACTION 0.198978 2.2 True
1554 1GJ6 X-RAY DIFFRACTION 0.0852393 1.5 True
1553 1GHZ X-RAY DIFFRACTION 0.0799989 1.39 True
1555 4BAH X-RAY DIFFRACTION 0.128 1.94 True
1556 3DA9 X-RAY DIFFRACTION 0.1187 1.8 True
1558 3P17 X-RAY DIFFRACTION 0.0531176 1.43 True
1559 3QWC X-RAY DIFFRACTION 0.0905379 1.75 True
1557 3RML X-RAY DIFFRACTION 0.0683916 1.53 True
Download load_all_experiments.py
Sites¶
Once we have uploaded the experiments, we can upload one or more Sites (OEDesignUnits) associated with each. Note, we could download the BU we just uploaded, run it through Spruce to create the OEDesignUnit(s) and then upload. But since, we already created the OEDesignUnits above, we can just upload them.
The one case we’ll will see later is for apo sites. With no defined ligand, we will create apo sites as needed when adding them to a project (where the actual site location is known in the project reference structure)
$ mmdscli --profile local site add -h
Usage: mmdscli site add [OPTIONS] ExperimentID DU
Add a new site into MMDS
Options:
--bfactor-image PATH Grapheme SVG depicting bfactor
--interaction-image PATH Grapheme SVG depicting protein-ligand interactions
--density-image PATH Grapheme SVG depicting electron density overlap
-y, --yes Say yes to any yes/no questions. [default: False]
--json Return JSON instead of simple text on stdout.
-q, --quiet Minimal output
-v, --verbose Verbose output
-h, --help Show this message and exit.
If the SVGs have been created locally, they can be passed on the command line. If not, they will be created as part of the add command.
To upload the main site for 1G3D, we need to associate this site with the Experiment already uploaded. We can get the ID and then upload the Site.
$ mmdscli --profile local experiment list --code 1G3D --fields id,code
id code public
---- ------ --------
1550 1G3D True
$ mmdscli --profile local site add 1550 trypsin/1G3D_A__DU__108_A-601.oedu --json
{
"bfactor_image": null,
"components": {
"cofactors": null,
"counter_ions": null,
"excipients": {
"json": "http://localhost:8080/api/v1/site/1449/excipients/",
"mask": "512",
"title": "excipients",
"url": "http://localhost:8080/api/v1/site/1449/excipients/1G3D_A-108_A-601_excipients.oeb"
},
"ligand": {
"json": "http://localhost:8080/api/v1/site/1449/ligand/",
"mask": "ligand",
"title": "108(A-601)",
"url": "http://localhost:8080/api/v1/site/1449/ligand/1G3D_A-108_A-601_ligand.oeb"
},
"lipids": null,
"metals": {
"json": "http://localhost:8080/api/v1/site/1449/metals/",
"mask": "metals",
"title": "metals",
"url": "http://localhost:8080/api/v1/site/1449/metals/1G3D_A-108_A-601_metals.oeb"
},
"nucleic": null,
"other_cofactors": null,
"other_ligands": null,
"other_nucleics": null,
"other_proteins": null,
"packing_residues": {
"json": "http://localhost:8080/api/v1/site/1449/packing_residues/",
"mask": "packing_residues",
"title": "packing residues",
"url": "http://localhost:8080/api/v1/site/1449/packing_residues/1G3D_A-108_A-601_packing_residues.oeb"
},
"protein": {
"json": "http://localhost:8080/api/v1/site/1449/protein/",
"mask": "protein",
"title": "1G3D(A)",
"url": "http://localhost:8080/api/v1/site/1449/protein/1G3D_A-108_A-601_protein.oeb"
},
"solvent": {
"json": "http://localhost:8080/api/v1/site/1449/solvent/",
"mask": "solvent",
"title": "solvent",
"url": "http://localhost:8080/api/v1/site/1449/solvent/1G3D_A-108_A-601_solvent.oeb"
},
"sugars": null
},
"density_image": {
"pk": 2462,
"url": "http://localhost:8080/api/mmds/images/2462/1G3D_A-108_A-601_density.svg"
},
"design_unit": {
"filename": "1G3D_A__DU__108_A-601.oedu",
"json": "http://localhost:8080/api/v1/site/1449/design_unit/",
"pk": 1449,
"url": "http://localhost:8080/api/v1/site/1449/design_unit/1G3D_A__DU__108_A-601.oedu"
},
"experiment": {
"DPI": 0.0897862915057751,
"RFree": 0.189,
"RFree_found": false,
"author": "E.TOYOTA,K.K.S.NG,H.SEKIZAKI,K.ITOH,K.TANIZAWA,M.N.G.JAMES",
"biounit": [
{
"experiment": 1550,
"filename": "1G3D_A__DU__biounit.oedu",
"id": 1744,
"url": "http://localhost:8080/api/v1/biounit/1744/1G3D_A__DU__biounit.oedu"
}
],
"code": "1G3D",
"created": "2019-06-23T17:28:51.872454Z",
"density": {
"filename": "1g3d.mtz",
"json": null,
"pk": 1169,
"url": "http://localhost:8080/api/v1/experiment/1550/density/1g3d.mtz"
},
"id": 1550,
"keywords": "ENZYME-INHIBITOR COMPLEX, COORDINATION METAL BASED INHIBITOR,HYDROLASE",
"ligands": [
{
"Id": "108",
"Smiles": "C[C@@H](C(=O)O)NCc1cc(ccc1O)C(=N)N",
"Tautomers": [],
"Title": "108",
"Type": "unknown"
}
],
"method": "X-RAY DIFFRACTION",
"public": true,
"resolution": 1.8,
"revision": "1.3",
"revision_date": "2018-04-04T00:00:00Z",
"sites": [
1449
],
"solved_date": "2001-01-17T00:00:00Z",
"strdate": "17-JAN-01",
"structure": {
"filename": "1G3D.pdb",
"json": "http://localhost:8080/api/v1/experiment/1550/structure/",
"pk": 5973,
"url": "http://localhost:8080/api/v1/experiment/1550/structure/1G3D.pdb"
},
"tags": []
},
"favorite": 1000,
"id": 1449,
"interaction_image": {
"pk": 2461,
"url": "http://localhost:8080/api/mmds/images/2461/1G3D_A-108_A-601_interactions.svg"
},
"iridium_score": "MT",
"ligand": {
"id": 568,
"ligandID": "108",
"smiles": "C[C@@H](C(=O)[O-])NCc1cc(ccc1[O-])C(=[NH2+])N",
"tags": []
},
"ligand_image": {
"data": "http://localhost:8080/depict/?smiles=C%5BC%40%40H%5D%28C%28%3DO%29%5BO-%5D%29NCc1cc%28ccc1%5BO-%5D%29C%28%3D%5BNH2%2B%5D%29N",
"pk": -1
},
"ligand_title": "108(A-601)",
"surface_file": {
"filename": "1G3D_A-108_A-601_surface.oesrf",
"json": "http://localhost:8080/api/v1/site/1449/surface/",
"pk": 5983,
"url": "http://localhost:8080/api/v1/site/1449/surface/1G3D_A-108_A-601_surface.oesrf"
},
"tags": [],
"title": "1G3D(A) > 108(A-601)"
}
$ mmdscli --profile local status
version oe_license_expires families experiments sites contexts
--------- -------------------- ---------- ------------- ------- ----------
1.0.0 2019-09-04 0 10 1 0
Note that this is just the original site for 1G3D. The power of MMDS is associating groups of sites into projects (contexts) with an associated superposition method.
Just like Experiments before, we can continue through each directory loading sites one at a time, or we can use a script that uses the mmdsclient Python API to load all the sites in a single directory at once.
Note
Since these are simple Python scripts, the MMDS_PROFILE environment variable needs to be set in order for the mmdsclient API to load the appropriate credentials.
$ MMDS_PROFILE=local python load_all_sites.py trypsin
✔ Gathering existing sites
✔ Gathering input DU files : 5
processing 5 groups of OEDesignUnit files [*****************************] 100.0%
Elapsed: 5.72 secs
$ MMDS_PROFILE=local python load_all_sites.py thrombin
✔ Gathering existing sites : 6
✔ Gathering input DU files : 5
processing 5 groups of OEDesignUnit files [*****************************] 100.0%
Elapsed: 5.64 secs
And a quick check:
Warning
Note that for 1000+ sites, this command will take some time, so be careful.
$ mmdscli --profile local site list
id title iridium_score favorite ligand_title
---- ----------------------- --------------- ---------- --------------
1460 1TRN(A) > apo(A-226) NT 1000 apo(A-226)
1461 1G3D(A) > 108(A-601) MT 1000 108(A-601)
1462 1TRN(B) > apo(B-226) NT 1000 apo(B-226)
1463 1K1L(A) > FD3(A-999) MT 1000 FD3(A-999)
1464 1GHZ(A) > 120(A-246) MT 1000 120(A-246)
1465 1GJ6(A) > 132(A-246) MT 1000 132(A-246)
1466 3P17(HIL) > 99P(H-1001) MT 1000 99P(H-1001)
1467 3QWC(HIL) > 98P(H-2001) MT 1000 98P(H-2001)
1468 3DA9(ABD) > 44U(B-1) HT 1000 44U(B-1)
1469 4BAH(ABD) > MEL(B-1291) MT 1000 MEL(B-1291)
1470 3RML(HIL) > M31(H-1) MT 1000 M31(H-1)
Download load_all_sites.py
Families¶
Family Tree¶
In order to organize structures into projects/contexts, we need to create a family tree containing the ultimate arrangement of structures by family and sub-family. We will collect structures by project, but these projects (contexts) are the leaves of the family tree.
The command mmds family can be used to add and manage this tree.
$ mmdscli family
Usage: mmdscli family [OPTIONS] COMMAND [ARGS]...
Manage MMDS families
Options:
-y, --yes Say yes to any yes/no questions. [default: False]
--json Return JSON instead of simple text on stdout.
-q, --quiet Minimal output
-v, --verbose Verbose output
-h, --help Show this message and exit.
Commands:
add Add a new family to MMDS
delete Delete one MMDS family
deleteall Delete all MMDS families
info Show info for one MMDS family
list List MMDS families
update Update a new family in MMDS
For families that are expected to be overlain in 3D, we will include a reference structure and a superposition method to use to align the sub-family/project reference structures. Spruce superposition methods include GlobalSequence, SiteSequence, DDM, and SSE.
For our example tree, the root will be RCSB, with the whole tree looking something like:
RCSB +
|
+ Protease +
|
+ Serine Protease
In this case, we want to provide a reference Serine Protease structure and method so that both of our contexts, Trypsin and Thrombin will be superposed into the same frame of reference. We will be using an OEDesignUnit from from 1G3D as the reference.
This set of commands will create the tree we want:
$ mmdscli --profile local family add RCSB
id title alignable unique_name
---- ------- ----------- -------------
1662 RCSB False RCSB
$ mmdscli --profile local family add Protease --parent RCSB
id title alignable parent unique_name
---- -------- ----------- -------- -------------
1663 Protease False 1662 RCSB|Protease
$ mmdscli --profile local family add "Serine Protease" --parent "RCSB|Protease" --reference trypsin/1G3D_A__DU__108_A-601.oedu
id title alignable method parent unique_name
---- --------------- ----------- ------------ -------- -----------------------------
1664 Serine Protease True SiteSequence 1663 RCSB|Protease|Serine Protease
$ mmdscli --profile local family list
id title alignable unique_name parent method
---- --------------- ----------- ----------------------------- -------- ------------
1662 RCSB False RCSB
1663 Protease False RCSB|Protease 1662
1664 Serine Protease True RCSB|Protease|Serine Protease 1663 SiteSequence
Contexts¶
We need to create a “Trypsin” context/project under the “Protease|Serine Protease” part of the family tree.
$ mmdscli --profile local status
version oe_license_expires families experiments sites contexts
---------- -------------------- ---------- ------------- ------- ----------
0.13.0dev1 2019-07-02 20 1 1 0
Contexts require a title and a parent Family. Note that the combo of title and parent Family title needs to be unique. So for this example, we will create “RCSB|Protease|Serine Protease|Trypsin”. The parent Family ID from above is 1664.
Contexts also require a reference OEDesignUnit (which designates the site of interest) and an appropriate superposition method. SiteSequence is recommended. Turns out 1G3D is a good reference DU so we can use it for this context.
$ mmdscli --profile local context add --title Trypsin --family 1664 --ref-structure trypsin/1G3D_A__DU__108_A-601.oedu --json
{
"active": true,
"context_frames": [],
"created": "2019-06-25T11:10:29.369075Z",
"family": {
"alignable": true,
"children": [],
"id": 1664,
"method": "SiteSequence",
"parent": 1663,
"reference": {
"filename": "1G3D_A__DU__108_A-601.oedu",
"json": "http://localhost:8080/api/v1/file/6011/",
"pk": 6011,
"url": "http://localhost:8080/api/v1/file/6011/1G3D_A__DU__108_A-601.oedu"
},
"title": "Serine Protease",
"unique_name": "RCSB|Protease|Serine Protease"
},
"id": 775,
"method": "SiteSequence",
"public": true,
"ref_structure": {
"filename": "1G3D_A__DU__108_A-601.oedu",
"json": "http://localhost:8080/api/v1/context/775/ref_structure/",
"pk": 6012,
"url": "http://localhost:8080/api/v1/context/775/ref_structure/1G3D_A__DU__108_A-601.oedu"
},
"title": "Trypsin",
"type": "project",
"updated": "2019-06-25T11:10:29.386860Z",
"view": null
}
$ mmdscli --profile local status
version oe_license_expires families experiments sites contexts
---------- -------------------- ---------- ------------- ------- ----------
0.13.0dev1 2019-07-02 20 1 1 1
Now do the same for Thrombin
$ mmdscli --profile local context add --title Thrombin --family 1664 --ref-structure thrombin/3RML_HIL__DU__M31_H-1.oedu --json
{
"active": true,
"context_frames": [],
"created": "2019-06-25T11:12:14.353287Z",
"family": {
"alignable": true,
"children": [],
"id": 1664,
"method": "SiteSequence",
"parent": 1663,
"reference": {
"filename": "1G3D_A__DU__108_A-601.oedu",
"json": "http://localhost:8080/api/v1/file/6011/",
"pk": 6011,
"url": "http://localhost:8080/api/v1/file/6011/1G3D_A__DU__108_A-601.oedu"
},
"title": "Serine Protease",
"unique_name": "RCSB|Protease|Serine Protease"
},
"id": 776,
"method": "SiteSequence",
"public": true,
"ref_structure": {
"filename": "3RML_HIL__DU__M31_H-1.oedu",
"json": "http://localhost:8080/api/v1/context/776/ref_structure/",
"pk": 6013,
"url": "http://localhost:8080/api/v1/context/776/ref_structure/3RML_HIL__DU__M31_H-1.oedu"
},
"title": "Thrombin",
"type": "project",
"updated": "2019-06-25T11:12:14.359154Z",
"view": null
}
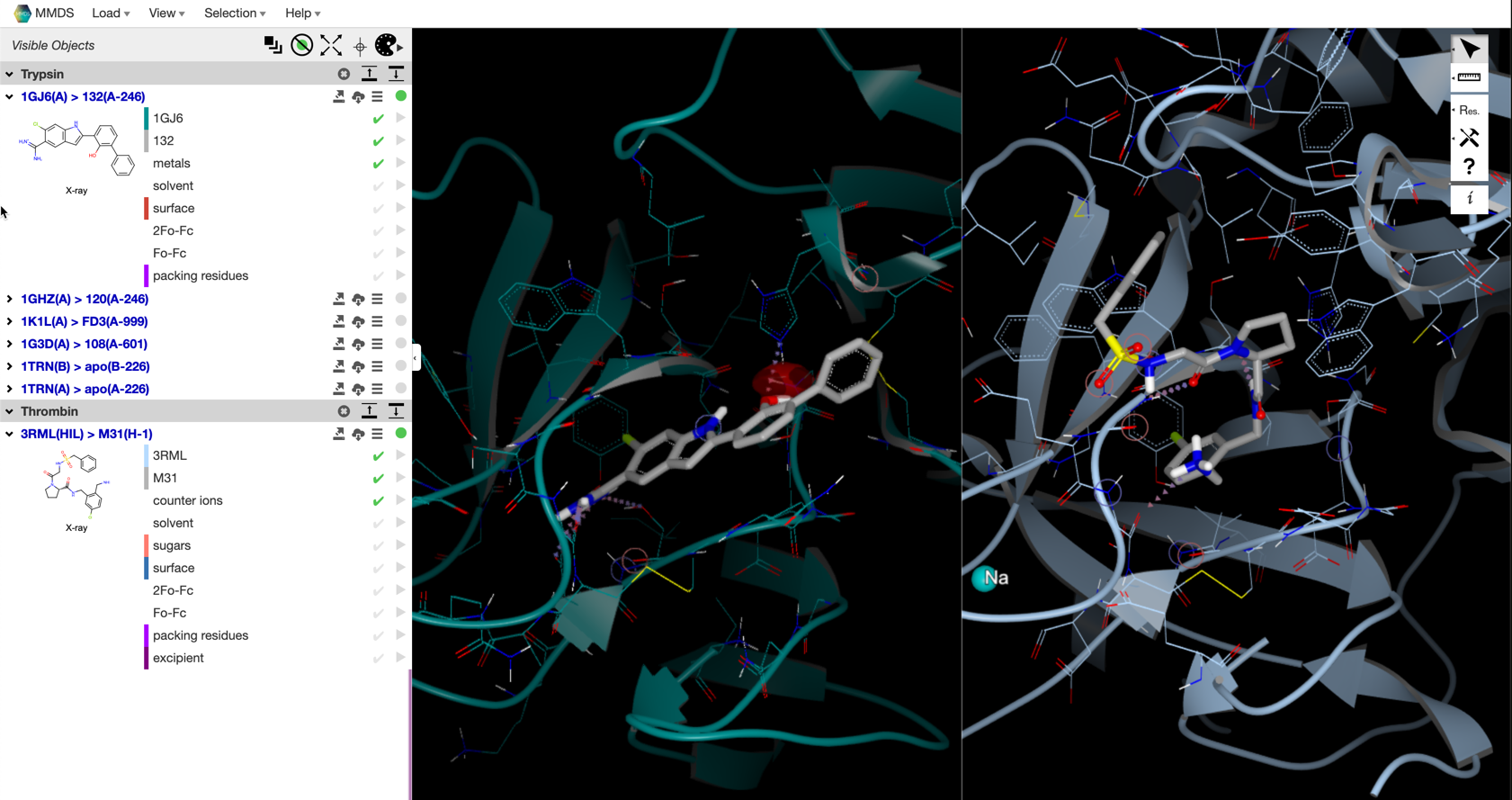
Frames¶
As Sites are added to Contexts, we create a new Frame object that holds a reference to the original Site and information about the 3D transform necessary to superpose into the current Context. As part of this process, we also download the MTZ or map file (if it exists) and create site-local grids for visualization.
$ mmdscli --profile local frame add -h
Usage: mmdscli frame add [OPTIONS] CONTEXT_ID SITE_ID
Add site/frame into an MMDS context
Options:
-y, --yes Say yes to any yes/no questions. [default: False]
--json Return JSON instead of simple text on stdout.
-q, --quiet Minimal output
-v, --verbose Verbose output
-h, --help Show this message and exit.
So, basically, all we need is the Trypsin Context ID we just created and the 1G3D Site ID.
$ mmdscli --profile local context list
id title active created method public type updated
---- -------- -------- --------------------------- ------------ -------- ------- ---------------------------
775 Trypsin True 2019-06-25T11:10:29.369075Z SiteSequence True project 2019-06-25T11:10:29.386860Z
776 Thrombin True 2019-06-25T11:12:14.353287Z SiteSequence True project 2019-06-25T11:12:14.359154Z
$ mmdscli --profile local site list
id title iridium_score favorite ligand_title
---- ----------------------- --------------- ---------- --------------
1471 1TRN(A) > apo(A-226) NT 1000 apo(A-226)
1472 1G3D(A) > 108(A-601) MT 1000 108(A-601)
1475 1TRN(B) > apo(B-226) NT 1000 apo(B-226)
1473 1K1L(A) > FD3(A-999) MT 1000 FD3(A-999)
1474 1GHZ(A) > 120(A-246) MT 1000 120(A-246)
1476 1GJ6(A) > 132(A-246) MT 1000 132(A-246)
1477 3DA9(ABD) > 44U(B-1) HT 1000 44U(B-1)
1479 3QWC(HIL) > 98P(H-2001) MT 1000 98P(H-2001)
1478 3P17(HIL) > 99P(H-1001) MT 1000 99P(H-1001)
1480 4BAH(ABD) > MEL(B-1291) MT 1000 MEL(B-1291)
1481 3RML(HIL) > M31(H-1) MT 1000 M31(H-1)
$ mmdscli --profile local frame add 775 1472
id title context context_title created fit_chain_order ligandID ligand_title proteinID
---- -------------------- --------- --------------- --------------------------- ----------------- ---------- -------------- -----------
1245 1G3D(A) > 108(A-601) 775 Trypsin 2019-06-25T11:31:31.748126Z ['A'] 108 108(A-601) 1G3D
Adding Frames in bulk¶
$ MMDS_PROFILE=local python add_experiment_to_context.py 776 --dir thrombin
Adding 5 experiments to context id: 776
adding 3DA9
code: 3DA9 -> frame: 1256
adding 3P17
code: 3P17 -> frame: 1257
adding 4BAH
code: 4BAH -> frame: 1258
adding 3QWC
code: 3QWC -> frame: 1259
adding 3RML
code: 3RML -> frame: 1260
Do the same for the trypsin directory and Context 775:
$ MMDS_PROFILE=local python add_experiment_to_context.py 775 --dir trypsin
And finally we can see all the Frames we have loaded:
$ mmdscli --profile local frame list
id title context context_title created fit_chain_order ligandID ligand_title proteinID
---- ----------------------- --------- --------------- --------------------------- ----------------- ---------- -------------- -----------
1260 3RML(HIL) > M31(H-1) 776 Thrombin 2019-06-25T14:37:36.773416Z ['H', 'I', 'L'] M31 M31(H-1) 3RML
1245 1G3D(A) > 108(A-601) 775 Trypsin 2019-06-25T11:31:31.748126Z ['A'] 108 108(A-601) 1G3D
1246 1TRN(A) > apo(A-226) 775 Trypsin 2019-06-25T11:48:11.735570Z ['A'] apo apo(A-226) 1TRN
1247 1TRN(B) > apo(B-226) 775 Trypsin 2019-06-25T11:48:13.465583Z ['B'] apo apo(B-226) 1TRN
1248 1GJ6(A) > 132(A-246) 775 Trypsin 2019-06-25T11:58:41.762722Z ['A'] 132 132(A-246) 1GJ6
1249 1K1L(A) > FD3(A-999) 775 Trypsin 2019-06-25T11:58:44.550983Z ['A'] FD3 FD3(A-999) 1K1L
1250 1GHZ(A) > 120(A-246) 775 Trypsin 2019-06-25T11:58:48.019884Z ['A'] 120 120(A-246) 1GHZ
1256 3DA9(ABD) > 44U(B-1) 776 Thrombin 2019-06-25T14:37:22.831725Z ['B', 'D', 'A'] 44U 44U(B-1) 3DA9
1257 3P17(HIL) > 99P(H-1001) 776 Thrombin 2019-06-25T14:37:26.588846Z ['H', 'I', 'L'] 99P 99P(H-1001) 3P17
1258 4BAH(ABD) > MEL(B-1291) 776 Thrombin 2019-06-25T14:37:29.891082Z ['B', 'D', 'A'] MEL MEL(B-1291) 4BAH
1259 3QWC(HIL) > 98P(H-2001) 776 Thrombin 2019-06-25T14:37:33.119753Z ['H', 'I', 'L'] 98P 98P(H-2001) 3QWC
Download add_experiment_to_context.py