Fingerprint Coverage¶
Fingerprints are usually generated by enumerating various fragments (patterns)
of a molecule and then hashing them into a fixed-length bitvector.
The OEGetFPCoverage function provides access to these
fragments by returning an iterator over OEAtomBondSet objects,
each of which storing the atoms and bonds of a specific fragment.
See also
Fingerprint Patterns chapter that shows how to access more information about the fragments enumerated during the fingerprint generation
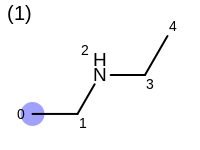
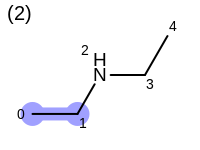
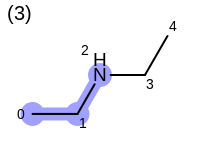
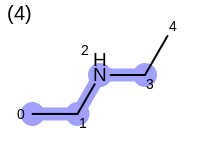
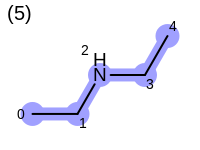
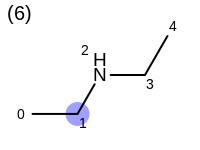
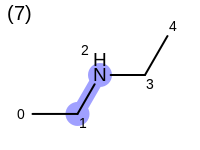
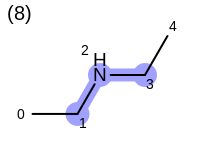
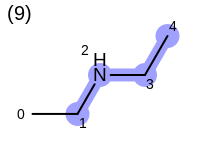
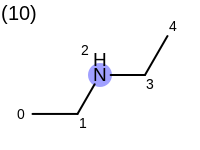
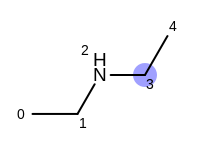
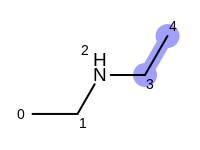
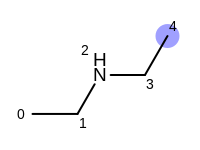
The following example shows how the retrieve the unique fragments that are enumerated when generating a path fingerprint. The obtained fragments are depicted in Table: Example of path fragments
Listing 17: Example of accessing patterns encoded into a fingerprint
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "CCNCC")
fptype = oegraphsim.OEGetFPType(oegraphsim.OEFPType_Path)
unique = True
for idx, abset in enumerate(oegraphsim.OEGetFPCoverage(mol, fptype, unique)):
print("%2d %s" % ((idx + 1), "".join([str(a) for a in abset.GetAtoms()])))
The output of Listing 17 is the following:
1 0 C
2 0 C 1 C
3 0 C 1 C 2 N
4 0 C 1 C 2 N 3 C
5 0 C 1 C 2 N 3 C 4 C
6 1 C
7 1 C 2 N
8 1 C 2 N 3 C
9 1 C 2 N 3 C 4 C
10 2 N
|
|
|
|
|
|
|
|
|
|
The OEGetFPCoverage function in the
Listing 17 example is called with a unique options.
This means that it returns only unique fragments, where a fragment
(i.e. subgraph) is considered unique, if it differs from all other
subgraphs identified previously by at least one atom or bond.
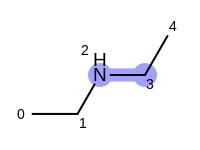
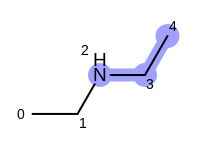
For example, executing the same code with a non-unique option would
generate five additional paths depicted in
Table: Example of additional non-unique path fragments
|
|
|
|
|
See also
OEGetFPOverlapfunctionFingerprint Overlap chapter