Fingerprint Overlap¶
The OEGetFPOverlap function provides access to the
fragments of two molecules that are considered equivalent based on a specific
fingerprint type.
This means that the returned fragment-pairs set the same bit “on” when
fingerprints are generated.
The following example shows how to retrieve the common five bond-length
patterns of two molecules.
Listing 18: Example of accessing common patterns based on a fingerprint
pmol = oechem.OEGraphMol()
oechem.OESmilesToMol(pmol, "c1cnc2c(c1)CC(CC2O)CF")
tmol = oechem.OEGraphMol()
oechem.OESmilesToMol(tmol, "c1cc2c(cc1)CC(CCl)CC2N")
fptype = oegraphsim.OEGetFPType("Tree,ver=2.0.0,size=4096,bonds=5-5,"
"atype=AtmNum|HvyDeg|EqHalo,btype=Order")
for idx, match in enumerate(oegraphsim.OEGetFPOverlap(pmol, tmol, fptype)):
ostring = "match %2d: " % (idx + 1)
for mpair in match.GetAtoms():
p = mpair.pattern
t = mpair.target
ostring += "%d%s-%d%s " % (p.GetIdx(), oechem.OEGetAtomicSymbol(p.GetAtomicNum()),
t.GetIdx(), oechem.OEGetAtomicSymbol(t.GetAtomicNum()))
print(ostring)
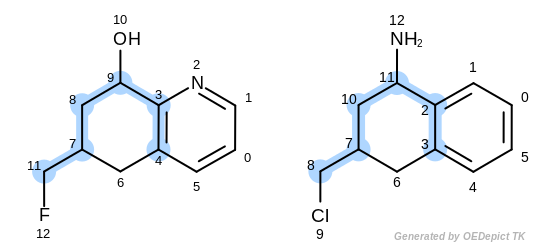
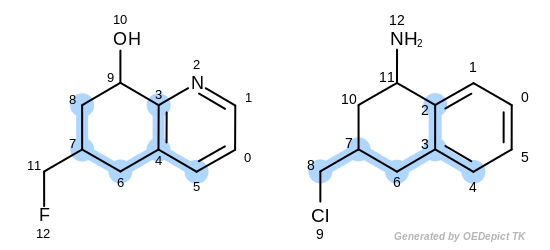
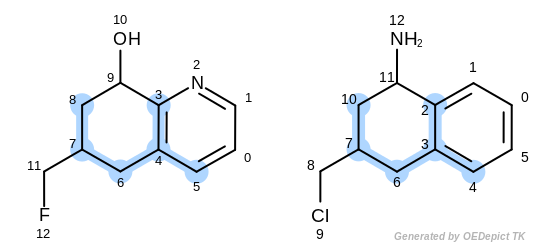
The first three matches returned by the Listing 18 are
depicted in the next table.
The output of code is the following:
match 1: 3C-2C 9C-11C 4C-3C 8C-10C 7C-7C 11C-8C
match 2: 3C-2C 4C-3C 5C-4C 6C-6C 7C-7C 8C-8C
match 3: 3C-2C 4C-3C 5C-4C 6C-6C 7C-7C 8C-10C
match 4: 3C-2C 4C-3C 5C-4C 6C-6C 7C-7C 11C-8C
match 5: 3C-2C 4C-3C 5C-4C 6C-6C 7C-7C 11C-10C
match 6: 3C-2C 9C-11C 4C-3C 6C-6C 7C-7C 11C-8C
... truncated ...
|
|
|
Warning
Even though the OEGetFPOverlap function returns
an iterator of OEMatchBase objects, they are not
matches in the traditional sense, i.e. the atom-pair and bond-pair
correspondences between the pattern and the target atoms and
bonds are not guaranteed. See example depicted below.
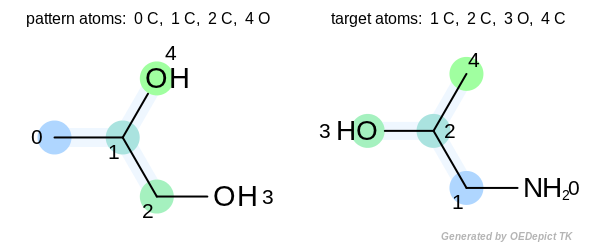
The two highlighted patterns set the same bit when fingerprints are generated, but the returned match is not pairwise. The numbers depicted next to the atoms are the atom indices.¶
See also
OEGetFPCoveragefunctionFingerprint Coverage chapter
Hint
The OEGetFPOverlap can be used to visualize molecule similarity based on
a given fingerprint type. See more details in: