Gaussian QM Run Input Files Successful Calculation¶
Running this floe with the provided input file will cost around $0.40.
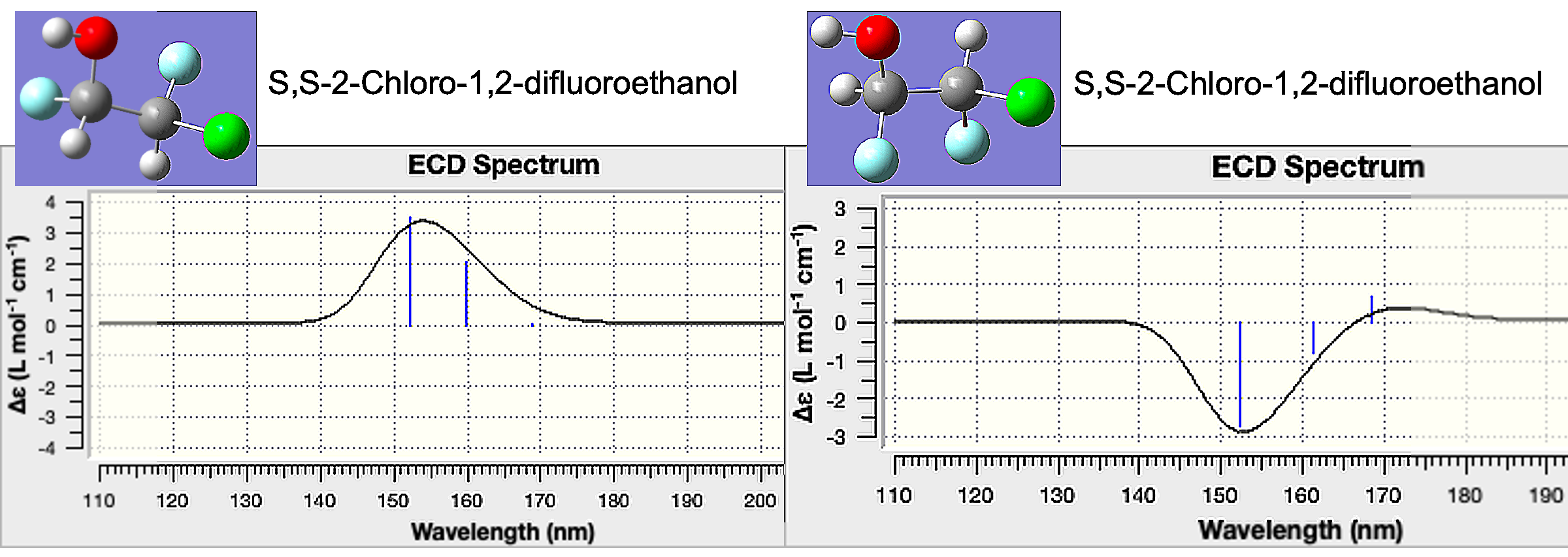
The input file for this floe (enantiomers.tar) can be found in the Gaussian QM Run Input Files folder in the Tutorial Data on Orion. This tar file has two Gaussian input files for 2-Chloro-1,2-difluoroethanol: rr_input.com and ss_input.com, corresponding to the R,R and S,S enantiomers, respectively. The output from this floe will let you compare the Electronic Circular Dichroism spectra (ECD) of the two enantiomers.
Locate and Run the Floe¶
Navigate through the categories Product-based / Quantum Mechanics / Gaussian to find this floe. Then locate or search for Gaussian QM Run Input Files.
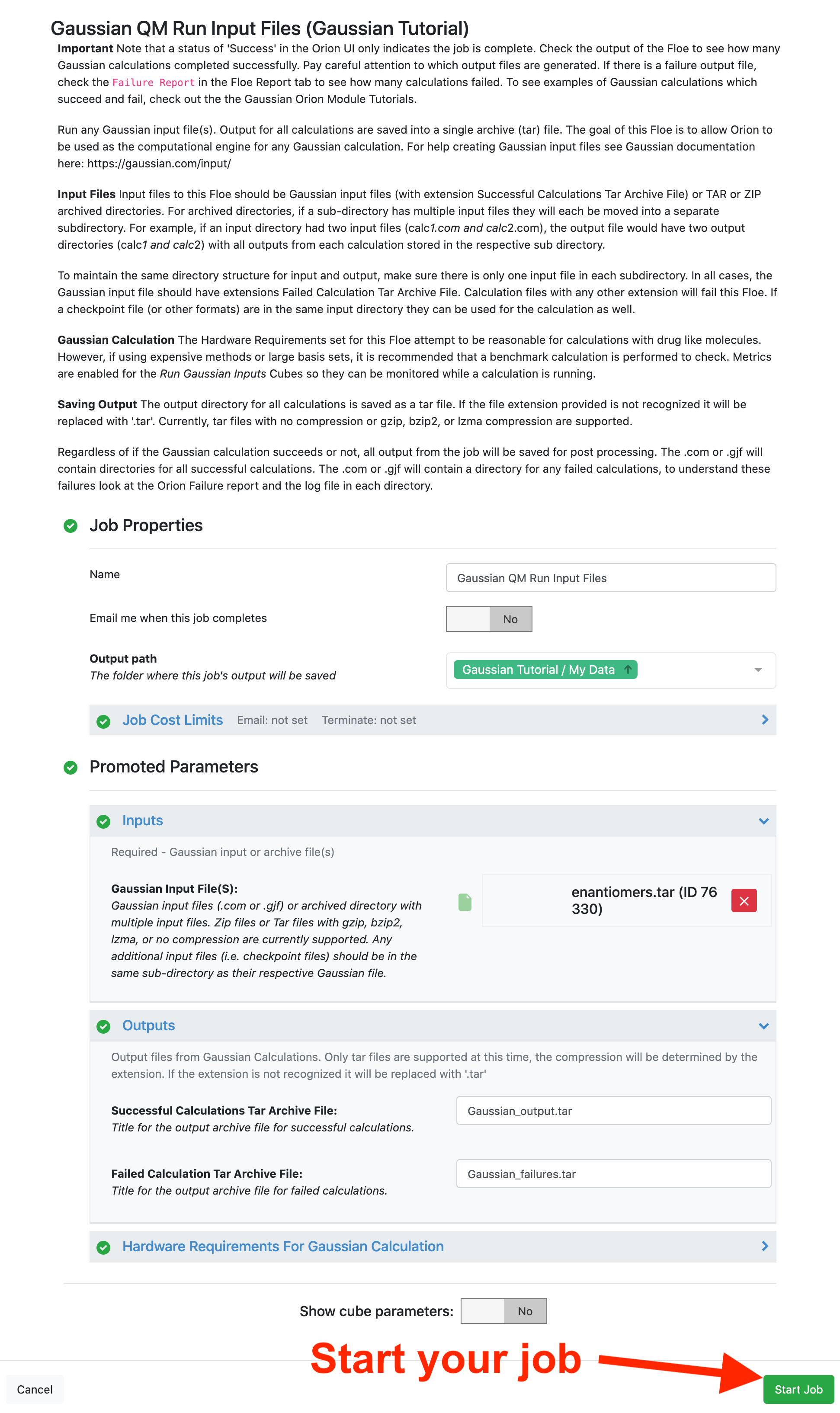
This floe takes any Gaussian files (extension .com or .gjf) or an archive with multiple Gaussian files as input (help organizing tar files). In this tutorial, the enantiomers.tar file from the Tutorial Data on Orion is used for the Gaussian Input Files parameter.
By default, the hardware requirements for Gaussian calculations are set to 8 CPUs, 14.4 GB memory, and 25.6 GB disk space. These parameters are meant to be more than sufficient for most drug-like molecules with most basis sets. However, for large calculations you may need to increase these requests. Press the “Start Job” button to begin the calculation:
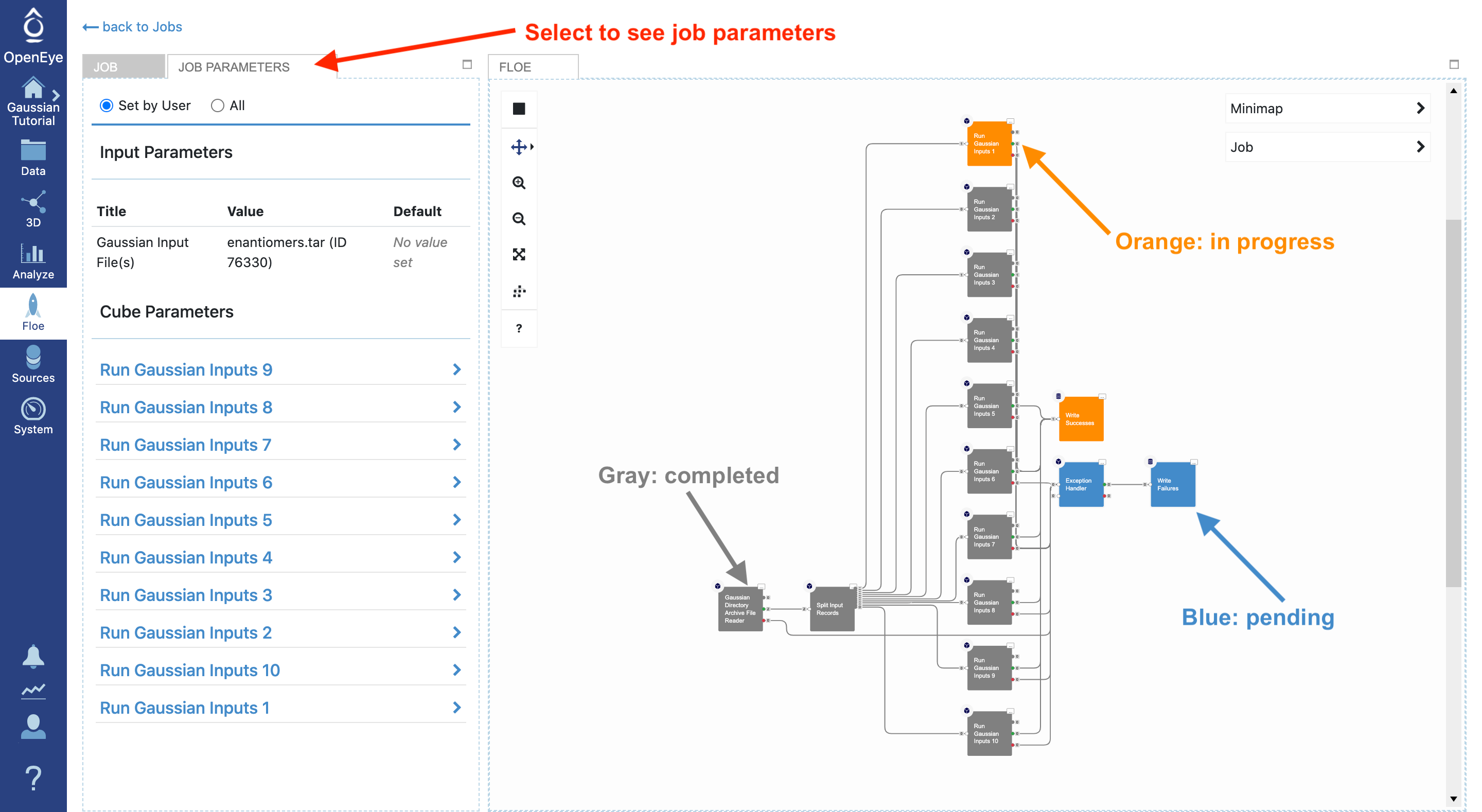
By navigating to the Jobs tab and selecting the running job, you can see the job details including the current cost and any written outputs. By selecting Job Parameters at the top, you can see changes to the parameters in this job compared to the floe default setting. The floe diagram on the right shows progress in the floe: gray, orange, and blue colors represent the completed, in progress, and pending tasks, respectively.
Download and Analyze Results¶
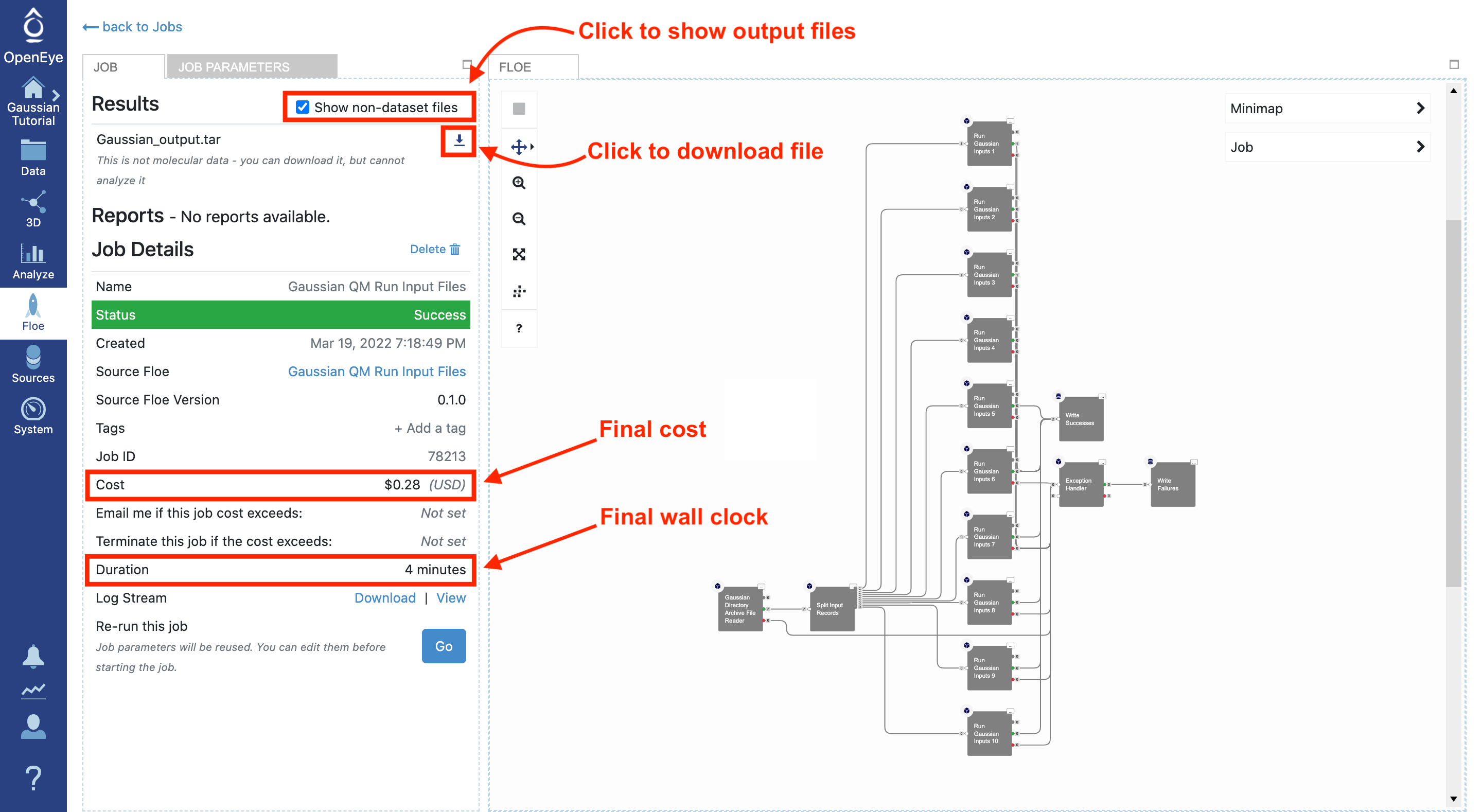
Once the job is finished, the job details will show the final cost and wall clock time. The generated outputs are shown in the Results section at the top left. This floe only generates files, so you must select the “Show non-dataset files” box to see the completed files. The results can be downloaded directly from here by clicking on the download arrow next to each file.
Note
Files can also be downloaded from the Data tab by clicking on the ellipses to the left of the file and choosing “Download.”
For this tutorial, there should only be one output file; we did not change the name when submitting the job, so it is called Gaussian_output.tar. If some or all of the calculations failed, there would be a Gaussian_failures.tar file. For an example with failure output, see the Gaussian QM Run Input Files Failed Calculation tutorial.
Download the Gaussian_output.tar file to your local machine.
Unpack the tar file by double clicking on the file or from the
command line with the command tar -xvf [path to file]
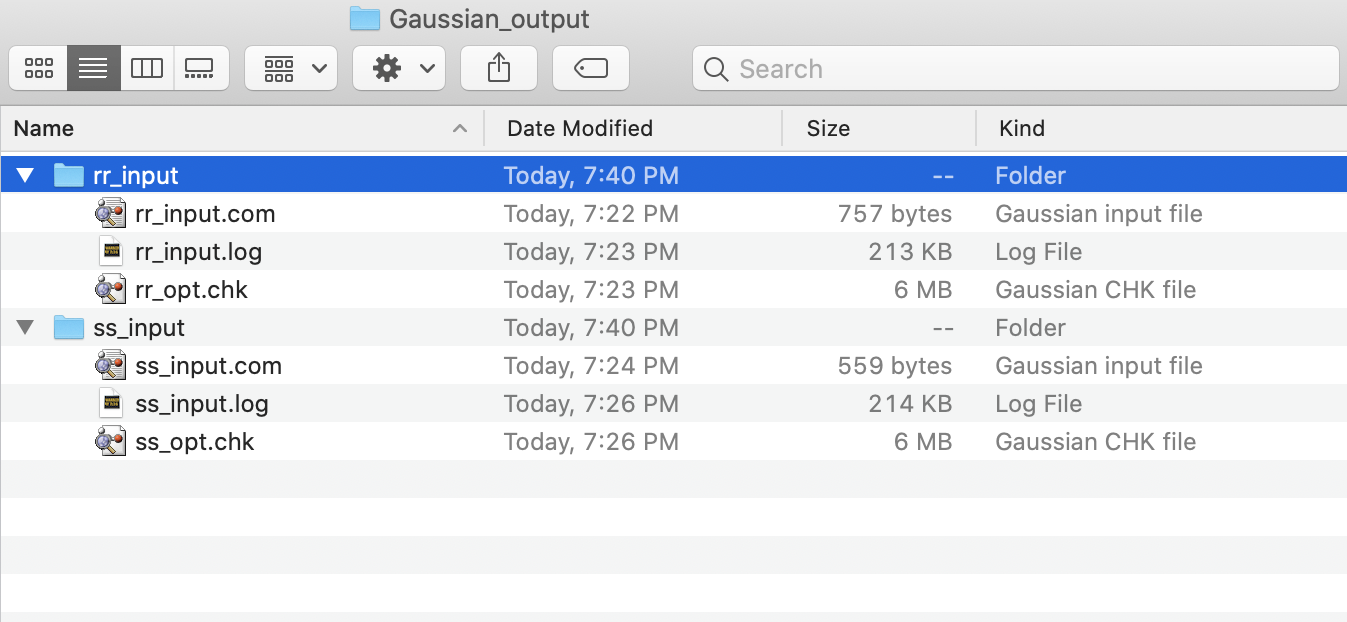
This will result in two directories, rr_input and ss_input.
Both will contain a copy of the input file (.com) and the resulting
output files (.log and .chk).
The results from this floe will always have the input file and a
log file. Any other result files must be specified in the
input file, such as the checkpoint files in this job.
Note
Currently, Gaussian_output.tar must be downloaded for analysis. In this tutorial, GaussView is used on a local machine to view the results. In the future, more integration with the Orion UI will be available.
Open the rr_opt.chk file with GaussView. Select Results > UV-Vis to show the UV-Vis and ECD spectra for this enantiomer.
Repeat the process above with the ss_opt.chk file. When we compare the ECD spectra, the two enantiomers have opposite changes in absorption. These can be compared to experimental results to understand the chiral purity of a sample.