Depicting Peptides
Problem
You want to depict a molecule as a peptide i.e. a series of amino acid components. See example in Figure 1.
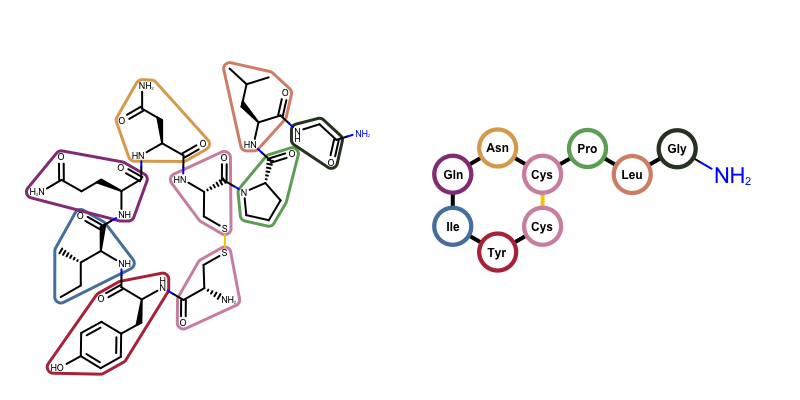
Figure 1. Example of depicting oxytocin as a molecule (on the left) and as a peptide (on the right)
Ingredients
|
Difficulty Level

Solution
The GraphSim TK provides a function called OEDrawPeptide that depicts a molecule as a peptide. It identifies standard amino acids components in the molecule and substitute them with circular glyphs and their corresponding code either 3-letter or single letter.
oechem.OEThrow.Fatal("Unknown image type!")
ofs = oechem.oeofstream()
if not ofs.open(oname):
oechem.OEThrow.Fatal("Cannot open output file!")
# read a molecules
mol = oechem.OEGraphMol()
Download code
peptide2img.py
and supporting data
oxytocin.ism
Usage:
prompt > python3 peptide2img.py oxytocin.ism oxytocin.png
Discussion
If svg image file format, the peptide can be depicted in interactive mode.
When the mouse is hovered over any glyph the corresponding amino acid is depicted.
prompt > python3 peptide2img.py -in oxytocin.ism -out oxytocin.svg -labelstyle threeletters
prompt > python3 peptide2img.py -in oxytocin.ism -out oxytocin.svg -labelstyle singleletter
Note
The generated interactive svg images should be included into and HTML page with the SVG MIME type.
<object data="<imagename>.svg" type="image/svg+xml"></object>
The amino acids that are appears naturally in proteins are the L-stereoisomers. These are labeled with corresponding 3-letter codes: such as Ser for L-Serine. Their enantiomers are distinguished by adding the “D-” prefix to their 3-letter code: such as D-Ser for D-Serine.
See also in OEDepict TK manual
Theory
Molecule Depiction chapter
API
OEImage class
OEWriteImage function
See also in GraphemeTM TK manual
API
OEDrawPeptide function
OEPeptideDisplayOptions class
OEPeptideLabelStyle namespace