Highlighting Fragments
Problem
You want to depict a molecule with highlighting of its fragments. See example in Table 1.
OEGetFuncGroupFragments |
OEGetRingChainFragments |
OEGetRingLinkerSideChainFragments |
Ingredients
|
Difficulty Level


Solution
The OEMedChem TK currently provides three ways to partition a molecule into fragments:
OEGetRingChainFragments - fragments a molecule into ring and chain components.
OEGetRingLinkerSideChainFragments - fragments a molecule into ring, linker and side-chain components as defined in [Bemis-1996] .
OEGetFuncGroupFragments - fragments a molecule into ring and functional group components.
The depict_molecule_with_fragment_highlights function, that takes the following parameters, shows how to depict molecules with fragment highlighting.
- image
An OEImage object which is a layout manager that allows generation of multi-page images.
- mol
The molecule being fragmented.
- fragfunc
The fragmentation function either OEGetRingChainFragments , OEGetRingLinkerSideChainFragments or OEGetFuncGroupFragments .
- opts
An OE2DMolDisplayOptions object that defines the style of the molecule depiction.
First, the molecule is fragmented by invoking the ‘fragfunc’ fragmentation function (line 3). The fragmentation function partitions the molecule and returns an iterator over OEAtomBondSet objects each storing the atoms and bonds of a fragment. An OELinearColorGradient object is then created, based on the number of returned fragments (lines 6-8), and is used to highlight each fragment with a distinct color. After constructing the molecule display, the fragments are looped over and highlighted using the OEAddHighlighting function.
1def depict_molecule_with_fragment_highlights(image, mol, fragfunc, opts):
2
3 fraglist = [f for f in fragfunc(mol)]
4 nrfrags = len(fraglist)
5
6 colorg = oechem.OELinearColorGradient()
7 colorg.AddStop(oechem.OEColorStop(0, oechem.OEMediumYellow))
8 colorg.AddStop(oechem.OEColorStop(nrfrags, oechem.OEDarkBrown))
9
10 disp = oedepict.OE2DMolDisplay(mol, opts)
11
12 highlight = oedepict.OEHighlightByLasso(oechem.OEWhite)
13 highlight.SetConsiderAtomLabelBoundingBox(True)
14
15 for fidx, frag in enumerate(fraglist):
16 highlight.SetColor(colorg.GetColorAt(fidx))
17 oedepict.OEAddHighlighting(disp, highlight, frag)
18
19 oedepict.OERenderMolecule(image, disp)
Download code
Usage:
prompt > python3 frags2img.py -in molecule.ism -out fragments.png -fragtype ring-chain
Discussion
The example below generates a multi-page PDF document.
At the top of each page an input molecule is rendered highlighting its fragments.
These fragments are then depicted one by one on the page.
See example in
Figure: Example of depiction of molecules with their fragments.
1def DepictMoleculesWithFragments(report, mollist, fragfunc,
2 moldispopts, fragdispopts):
3
4 for mol in mollist:
5
6 body = report.NewBody()
7 oedepict.OEPrepareDepiction(mol)
8 header = report.GetHeader(report.NumPages())
9
10 # loop over input molecule and fragment
11
12 fragsets = [f for f in fragfunc(mol)]
13 fragmols = []
14 for fset in fragsets:
15 fragment = oechem.OEGraphMol()
16 fragpred = oechem.OEIsAtomMember(fset.GetAtoms())
17 adjustHCount = True
18 oechem.OESubsetMol(fragment, mol, fragpred, adjustHCount)
19 fragmols.append(oechem.OEGraphMol(fragment))
20
21 nrfrags = len(fragmols)
22 colorg = oechem.OELinearColorGradient(oechem.OEColorStop(0, oechem.OEYellowTint),
23 oechem.OEColorStop(nrfrags - 1, oechem.OEDarkOrange))
24
25 # render molecule with fragment highlights
26
27 cellwidth, cellheight = report.GetHeaderWidth(), report.GetHeaderHeight()
28 moldispopts.SetDimensions(cellwidth, cellheight, oedepict.OEScale_AutoScale)
29
30 disp = oedepict.OE2DMolDisplay(mol, moldispopts)
31 for fidx, fset in enumerate(fragsets):
32 color = colorg.GetColorAt(fidx)
33 oedepict.OEAddHighlighting(disp, color, oedepict.OEHighlightStyle_BallAndStick, fset)
34
35 oedepict.OERenderMolecule(header, disp)
36
37 # create fragment grid
38
39 rows = max(2, int(math.sqrt(nrfrags + 1)))
40 cols = max(2, int(nrfrags / rows) + 1)
41 grid = oedepict.OEImageGrid(body, rows, cols)
42 grid.SetCellGap(8.0)
43
44 cellwidth, cellheight = grid.GetCellWidth(), grid.GetCellHeight()
45 fragdispopts.SetDimensions(cellwidth, cellheight, oedepict.OEScale_AutoScale)
46 fragdispopts.SetTitleLocation(oedepict.OETitleLocation_Hidden)
47
48 # determine the scale factor to depict fragments with equal size
49
50 minscale = oedepict.OEGetMoleculeScale(mol, fragdispopts) * 1.25
51 for frag in fragmols:
52 minscale = min(minscale, oedepict.OEGetMoleculeScale(frag, fragdispopts))
53 fragdispopts.SetScale(minscale)
54
55 # render each fragments
56
57 for fidx, (cell, fmol) in enumerate(zip(grid.GetCells(), fragmols)):
58 oedepict.OEPrepareDepiction(fmol)
59 disp = oedepict.OE2DMolDisplay(fmol, fragdispopts)
60 oedepict.OERenderMolecule(cell, disp)
61
62 color = colorg.GetColorAt(fidx)
63 pen = oedepict.OEPen(oechem.OEWhite, color, oedepict.OEFill_Off, 3.0)
64 oedepict.OEDrawBorder(cell, pen)
Download code
Usage:
prompt > python3 frags2pdf.py -in molecules.ism -out fragments.pdf -fragtype func-group
page 1 |
page 2 |
page 3 |
.. |
|---|---|---|---|
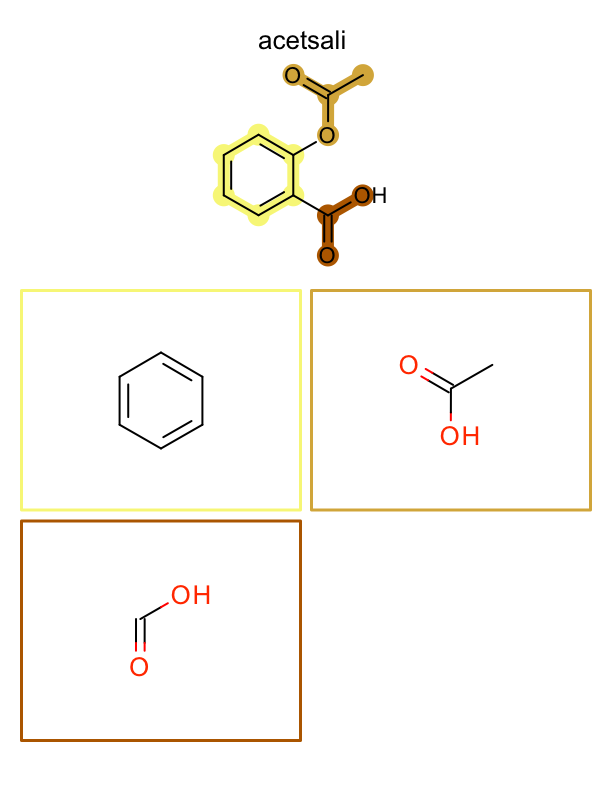
|
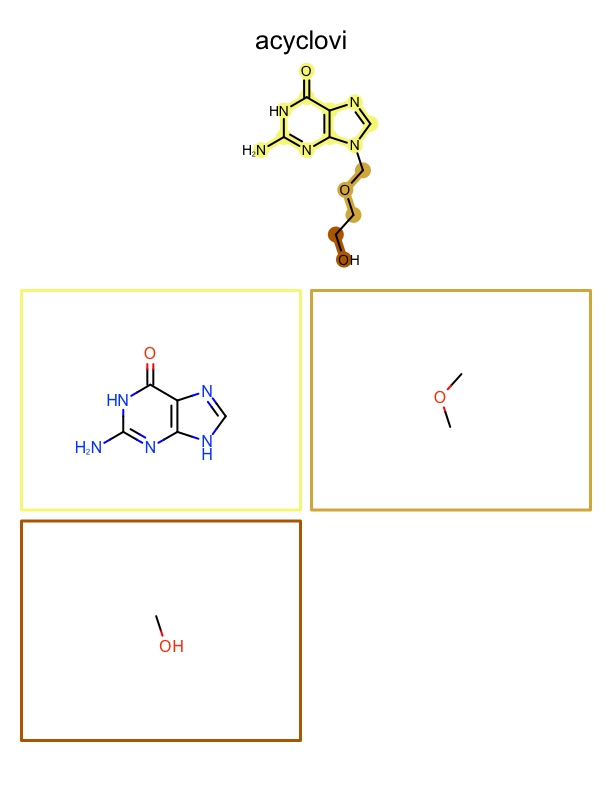
|
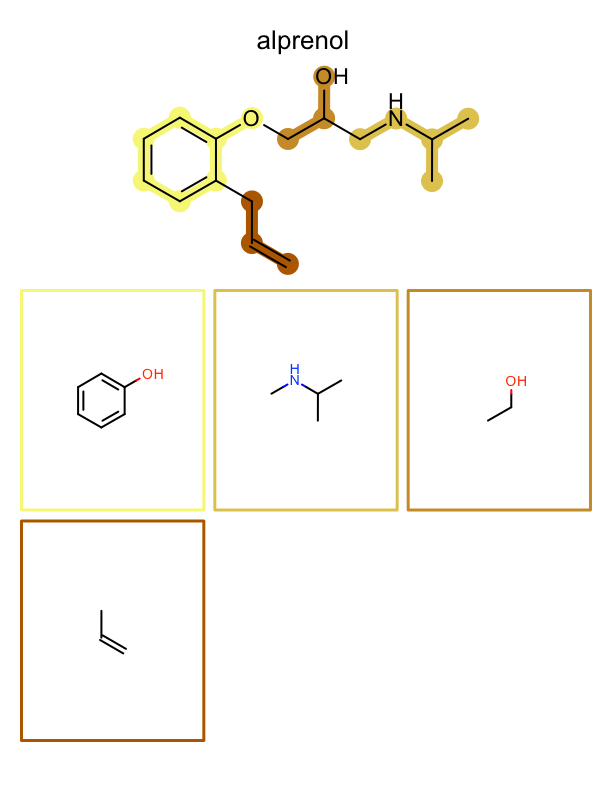
|
.. |
See also in OEChem TK manual
Theory
Predicates Functors chapter
API
OEAtomBondSet class
OEColorStop class
OEIsAtomMember predicate
OELinearColorGradient class
OESubsetMol function
See also in OEMedChem TK manual
Theory
Molecule Fragmentation chapter
API
OEGetFuncGroupFragments function
OEGetRingChainFragments function
OEGetRingLinkerSideChainFragments function
See also in OEDepict TK manual
Theory
Molecule Depiction chapter
Highlighting chapter
API
OE2DMolDisplay class
OE2DMolDisplayOptions class
OEAddHighlighting function
OEGetMoleculeScale function
OEImage class
OEImageGrid class
OEPrepareDepiction function
OERenderMolecule function