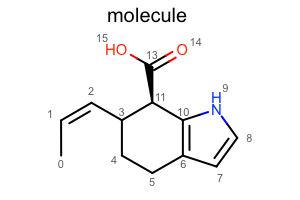
Depicting Molecule with Various Styles
Problem
You want to depict a molecule with various styles.
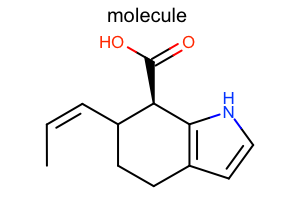
|
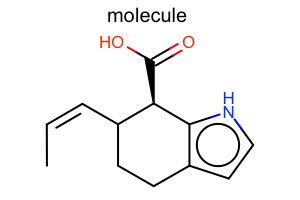
|
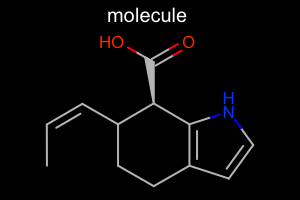
|
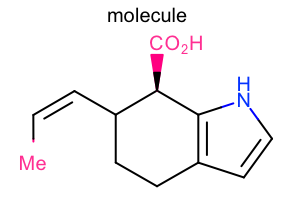
|
|
|
|
|
|
|
|
|
The examples below generate interactive .svg images
These svg images should be included into and HTML page with the SVG MIME type.
<object data="<imagename>.svg" type="image/svg+xml"></object>
|
|
|
|
Ingredients
|
Difficulty Level

Solution
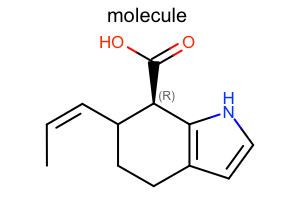
Default
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-default.png", disp)
Download code |

|
See also
OE2DMolDisplayOptions class
OE2DMolDisplay class
OERenderMolecule function
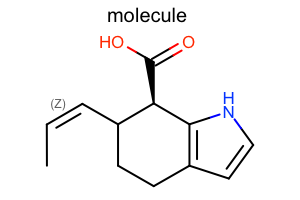
Aromaticity Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAromaticStyle(oedepict.OEAromaticStyle_Circle)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-aromatic-style.png", disp)
Download code |

|
See also
OEAromaticStyle namespace
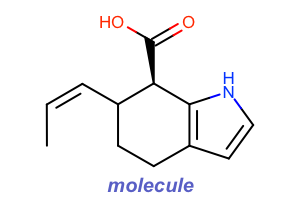
Atom Color Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomColorStyle(oedepict.OEAtomColorStyle_BlackCPK)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-color-style.png", disp)
Download code |

|
See also
OEAtomColorStyle namespace
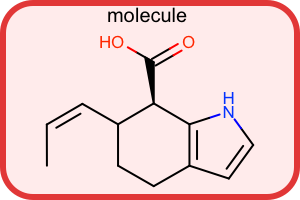
Super Atom Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetSuperAtomStyle(oedepict.OESuperAtomStyle_All)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-super-atom-style.png", disp)
Download code |

|
See also
OESuperAtomStyle namespace
Atom Stereo Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
stereostyle = oedepict.OEAtomStereoStyle_Display_All
stereostyle |= oedepict.OEAtomStereoStyle_HashWedgeStyle_Standard
opts.SetAtomStereoStyle(stereostyle)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-stereo-style.png", disp)
Download code |

|
See also
OEAtomStereoStyle namespace
Bond Stereo Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondStereoStyle(oedepict.OEBondStereoStyle_Display_All)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-stereo-style.png", disp)
Download code |

|
See also
OEBondStereoStyle namespace
Title Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetTitleLocation(oedepict.OETitleLocation_Bottom)
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold | oedepict.OEFontStyle_Italic
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OERoyalBlue)
opts.SetTitleFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-title.png", disp)
Download code |

|
See also
OETitleLocation namespace
OEFont class
Border Style
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
disp = oedepict.OE2DMolDisplay(mol, opts)
layer = disp.GetLayer(oedepict.OELayerPosition_Below)
lightred = oechem.OEColor(255, 235, 235) # R, G, B
mediumred = oechem.OEColor(225, 55, 55) # R, G, B
linewidth = 6.0
pen = oedepict.OEPen(lightred, mediumred, oedepict.OEFill_On, linewidth)
cornersize = 30
oedepict.OEDrawCurvedBorder(layer, pen, cornersize)
oedepict.OERenderMolecule("depiction-border.png", disp)
Download code |

|
See also
OE2DMolDisplay.GetLayer method
OELayerPosition namespace
OEPen class
OEDrawCurvedBorder method
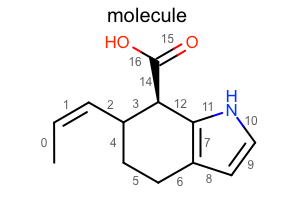
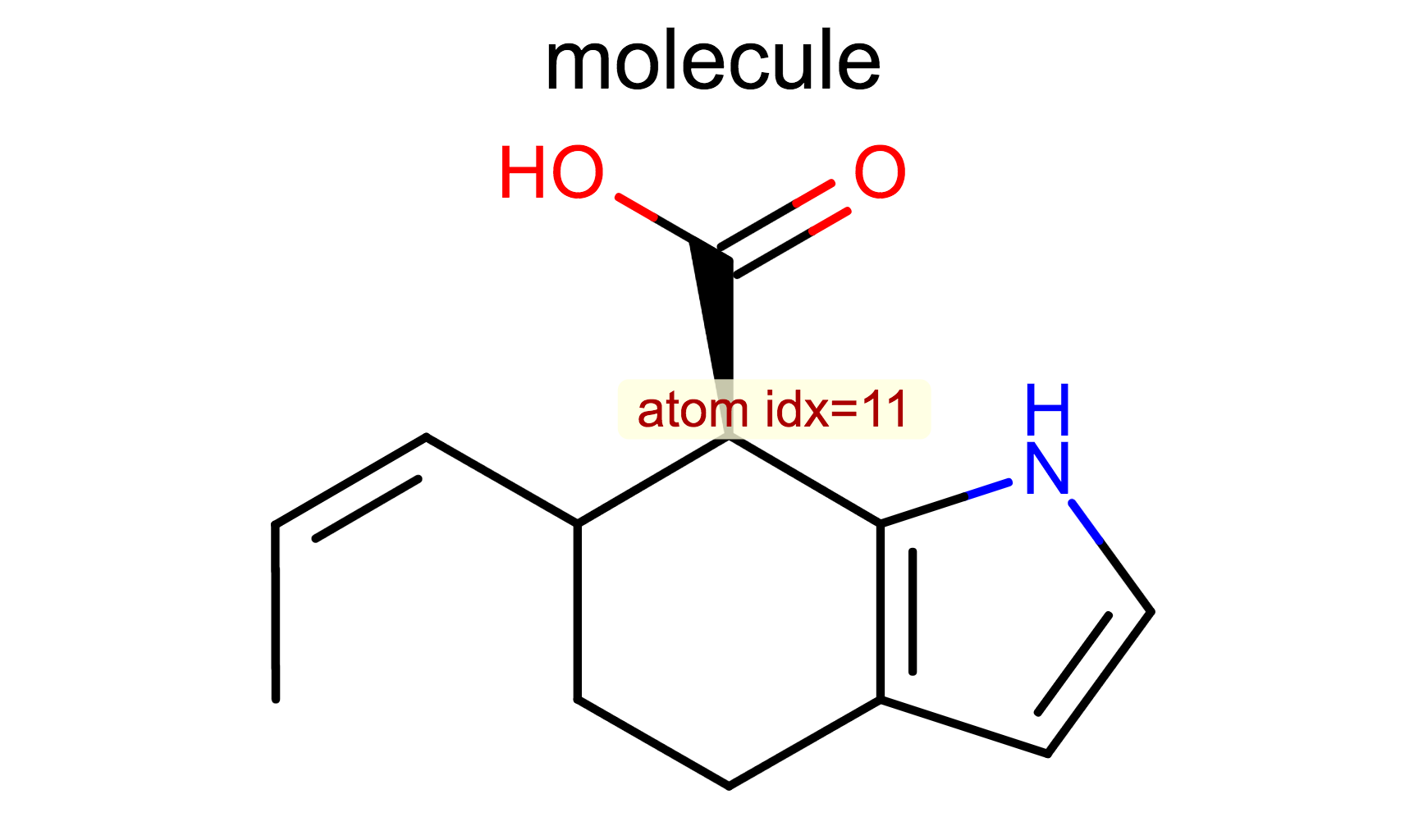
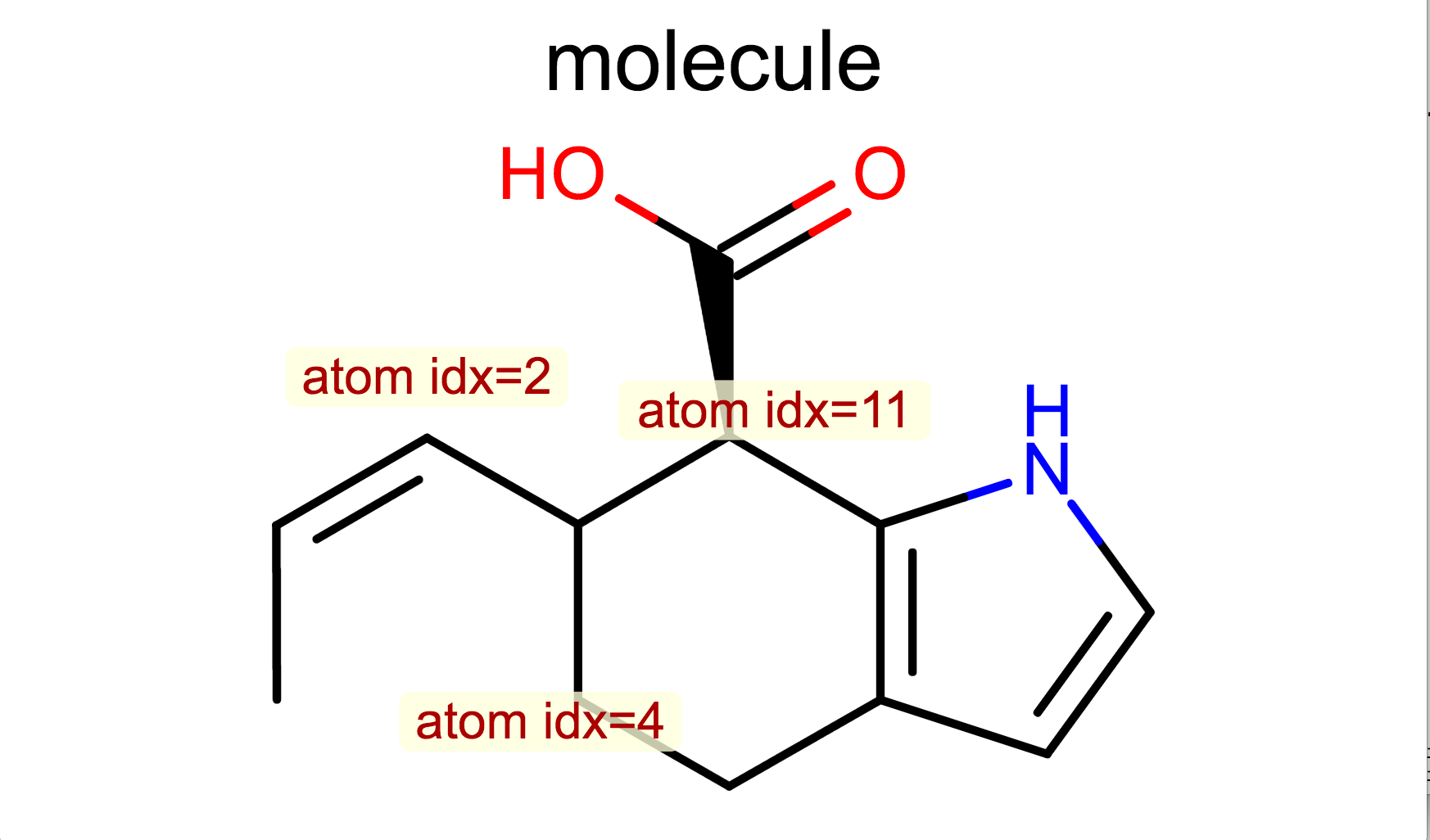
Display Atom Index
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomPropertyFunctor(oedepict.OEDisplayAtomIdx())
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-idx.png", disp)
Download code |

|
See also
OEDisplayAtomIdx class
OEDisplayAtomPropBase base class
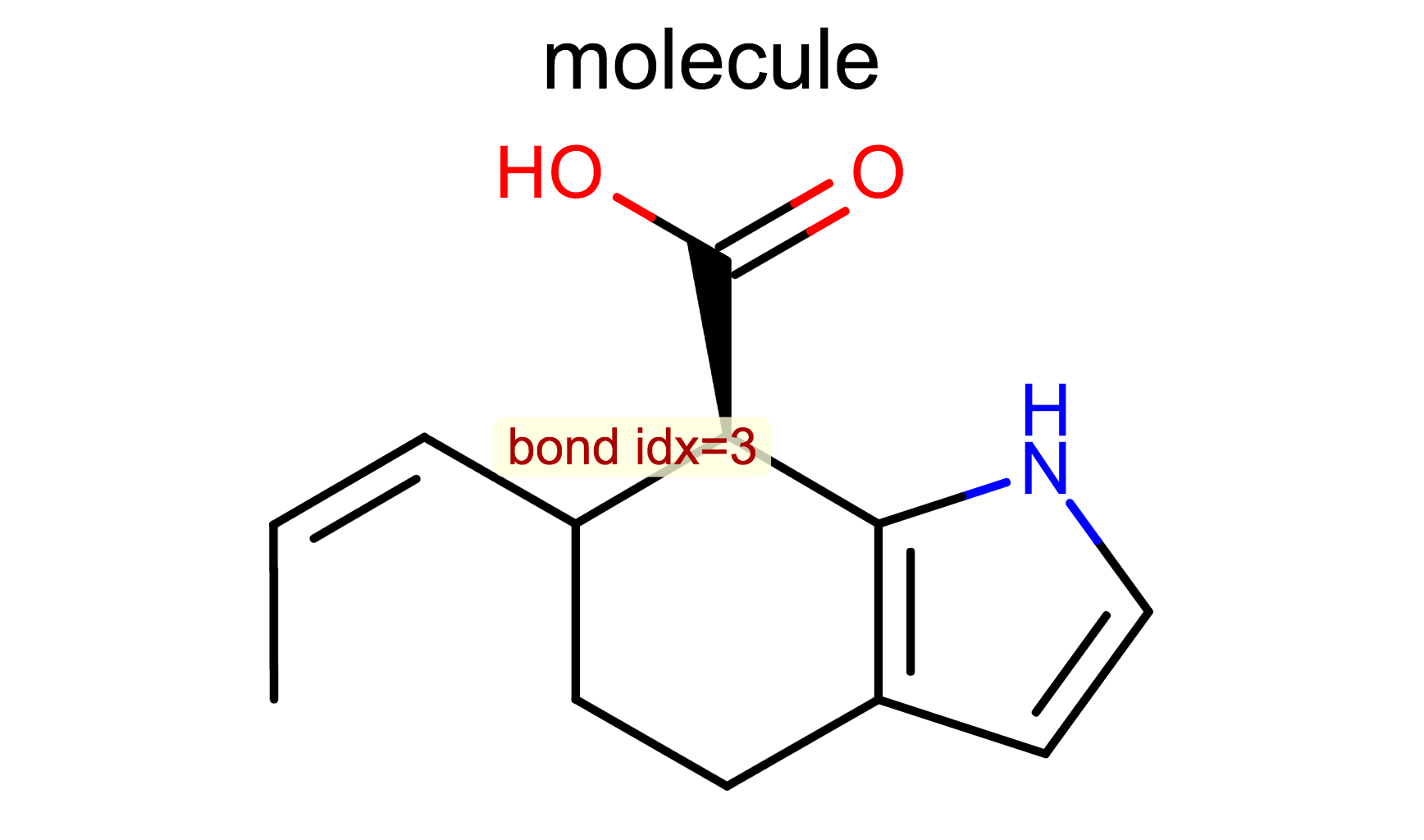
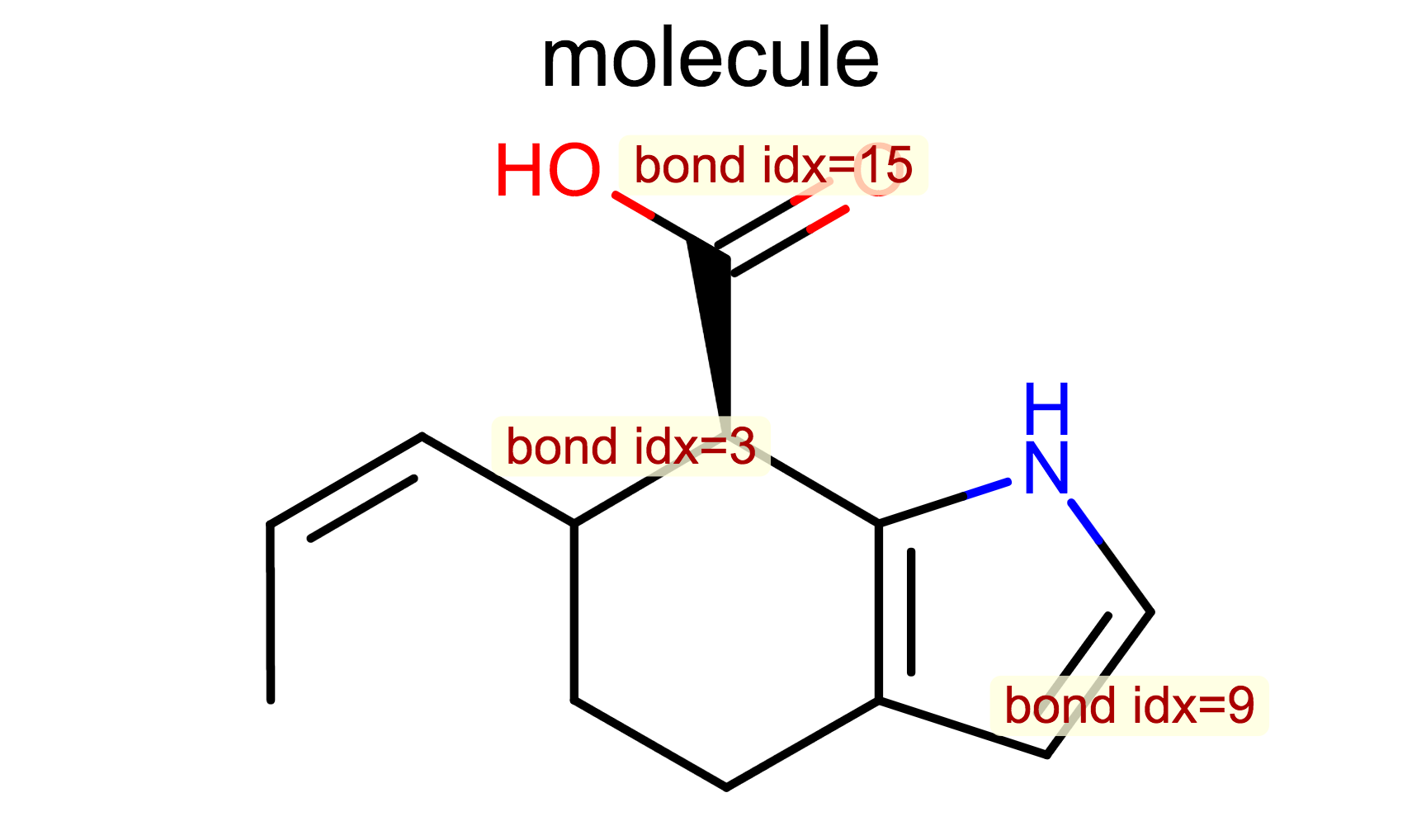
Display Bond Index
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondPropertyFunctor(oedepict.OEDisplayBondIdx())
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-idx.png", disp)
Download code |

|
See also
OEDisplayBondIdx class
OEDisplayBondPropBase base class
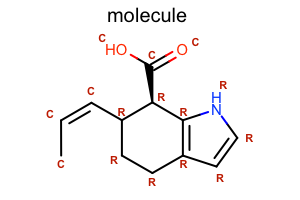
Display Atom Property
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
class LabelAtomTopology(oedepict.OEDisplayAtomPropBase):
def __init__(self):
oedepict.OEDisplayAtomPropBase.__init__(self)
def __call__(self, atom):
if atom.IsInRing():
return "R"
return "C"
def CreateCopy(self):
copy = LabelAtomTopology()
return copy.__disown__()
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomPropertyFunctor(LabelAtomTopology())
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OEDarkRed)
opts.SetAtomPropLabelFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-prop.png", disp)
Download code |

|
See also
OEDisplayAtomPropBase base class
OEFont class
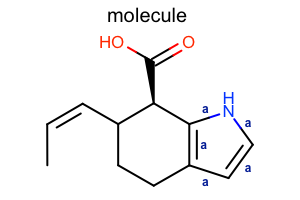
Display Bond Property
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
class LabelBondTopology(oedepict.OEDisplayBondPropBase):
def __init__(self):
oedepict.OEDisplayBondPropBase.__init__(self)
def __call__(self, bond):
if bond.IsAromatic():
return "a"
return ""
def CreateCopy(self):
copy = LabelBondTopology()
return copy.__disown__()
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondPropertyFunctor(LabelBondTopology())
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OEDarkBlue)
opts.SetBondPropLabelFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-prop.png", disp)
Download code |

|
See also
OEDisplayBondPropBase base class
OEFont class
Hover Atom Property
See also
OEFont class
OEDrawSVGHoverText function
Hover Bond Property
See also
OEFont class
OEDrawSVGHoverText function
Toggle Atom Property
See also
OEFont class
OEDrawSVGToggleText function
Toggle Bond Property
See also
OEFont class
OEDrawSVGToggleText function
See also in OEChem TK manual
API
OESmilesToMol function
See also in OEDepict TK manual
Theory
Molecule Depiction chapter
API
OE2DMolDisplay class
OE2DMolDisplayOptions class
OEPrepareDepiction function
OERenderMolecule function