Visualizing Protein-Ligand Unpaired Interactions
Problem
You want to visualize protein-ligand unpaired and clash interactions. See example in Figure 1.
Figure 1. Example of visualizing protein-ligand unpaired and clash interactions (PDB: 1NQ2))
Ingredients
|
Difficulty level


Download
Solution
The depict_unpairedmap illustrates how simple it is to generate these images.
OEInteractionHintContainer object is constructed that stores information about possible interactions between the ligand and the protein.
The interactions are perceived by calling the OEPerceiveInteractionHints function.
The active site is then prepared for 2D depiction by invoking the OEPrepareActiveSiteDepiction function.
When the OE2DActiveSiteDisplay object is constructed, residues are positioned around the ligand close to those atoms which they are interacting with.
The OERenderUnpairedInteractionMap function generates an image that displays the clash and unpaired interactions detected in the ligand and in nearby residues.
The legend associated with the unpaired map is rendered by invoking the OEDrawUnpairedInteractionMapLegend function.
1def depict_unpairedmap(image, protein, ligand, opts, legend_frame=None):
2 """
3 :type image: oedepict.OEImageBase
4 :type protein: oechem.OEMolBase
5 :type ligand: oechem.OEMolBase
6 :type opts: oedepict.OE2DMolDisplayOptions
7 :type legend_frame: oedepict.OEImageBase
8 """
9
10 # perceive interactions
11
12 asite = oechem.OEInteractionHintContainer(protein, ligand)
13 if not asite.IsValid():
14 oechem.OEThrow.Fatal("Cannot initialize active site!")
15 asite.SetTitle(ligand.GetTitle())
16
17 oechem.OEPerceiveInteractionHints(asite)
18
19 # depiction
20
21 oegrapheme.OEPrepareActiveSiteDepiction(asite)
22 adisp = oegrapheme.OE2DActiveSiteDisplay(asite, opts)
23 oegrapheme.OERenderUnpairedInteractionMap(image, adisp)
24
25 if legend_frame is not None:
26 lopts = oegrapheme.OE2DActiveSiteLegendDisplayOptions(12, 1)
27 oegrapheme.OEDrawUnpairedInteractionMapLegend(legend_frame, adisp, lopts)
Usage
Usage
The following commands will generate the image shown in Figure 1.
prompt > wget https://files.rcsb.org/download/1nq2.pdb
prompt > python3 unpairedmap2img.py -complex 1nq2.pdb -out 1nq2.svg -interactive-legend
Command Line Parameters
Simple parameter list
-height : Height of output image
-width : Width of output image
SplitMolComplex options :
-covalentligand : Split covalent ligands
-ligandname : Ligand name
molecule display options :
-aromstyle : Aromatic ring display style
input/output options :
-complex : Input filename of the protein-ligand complex
-protein : Input filename of the protein
-ligand : Input filename of the ligand
-out : Output filename of the generated image
active site display options:
-interactive-legend : Visualize legend on mouse hover (SVG feature)
Discussion
Interaction Perception
Currently the OEPerceiveInteractionHints function perceives the following interaction types:
Table 1. Interaction types currently available in OEChem TK name
corresponding interaction class
corresponding interaction type namespace
cation-pi
chelator
clash
None
contact
None
covalent
None
halogen bond
hydrogen bond
salt-bridge
stacking (T and Pi)
The default geometric parameters used by the OEPerceiveInteractionHints function have been set based on literature data ([Kumar-2002], [Cavallo-2016], [Bissantz-2010], and [Marcou-2007] ). The interaction parameters can be customized by using the OEPerceiveInteractionOptions class.
Unpaired Interaction Depiction
The OERenderUnpairedInteractionMap function currently visualizes the following interactions detected by the OEPerceiveInteractionHints function.
Atom clash interaction
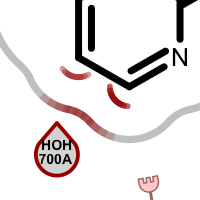
When visualizing protein-ligand atom crashes, red outline of a residue circle indicates that there are one or more atoms in that residue which are too close to some ligand atom(s). Clashing ligand atoms are marked with a red arc that is directed towards the corresponding clashing residue. The red shading on the grey line representing the shape of the pocket is used to identify atom clashes easily.
Table 2. Examples of visualizing atom clash(es) 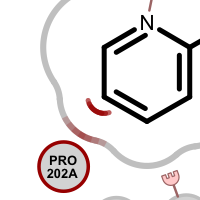
See also
OEClashInteractionHint class in the OEChem TK manual
Unpaired types of the hydrogen bonding interaction
An unpaired hydrogen bond interaction is detected:
if there is no ligand/protein acceptor atom that could interact with a protein/ligand donor atom
if there is no ligand/protein donor atom that could interact with a protein/ligand acceptor atom
Different linker types are used to mark unpaired acceptor and donor hydrogen bond interactions. Please note that since these interactions are unpaired, the direction of the linkers has no real spatial meaning. Ligand linkers are directed away from the ligand, while protein linkers are directed towards the ligand.
Table 3. Examples of visualizing unpaired hydrogen bond interactions 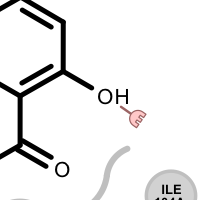
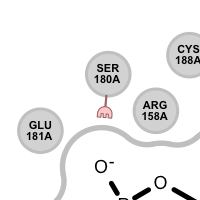
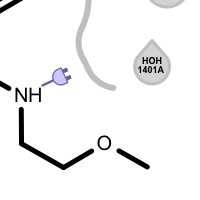
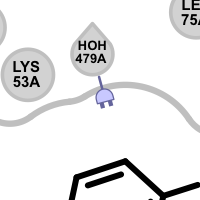
unpaired ligand acceptor
unpaired protein acceptor
unpaired ligand donor
unpaired protein donor
See also
OEHBondInteractionHint class in the OEChem TK manual
OEHBondInteractionHintType namespace in the OEChem TK manual
Clash types of the hydrogen bonding interaction
A hydrogen bond clash interaction is detected:
if an acceptor ligand atom interacts with an acceptor protein atom
if a donor ligand atom interacts with an donor protein atom
Table 4. Examples of visualization hydrogen bond clash interactions 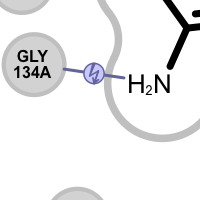
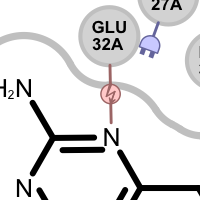
donor-donor clash
acceptor-acceptor clash
See also
OEHBondInteractionHint class in the OEChem TK manual
OEHBondInteractionHintType namespace in the OEChem TK manual
Unpaired types of the salt-bridge interaction
An unpaired salt bridge interaction is detected if there is a positively / negatively charged functional group either in the ligand or in nearby protein without a matching negatively / positively charged functional group, respectively.
Different linker types are used to mark unpaired positive and negative salt-bridge interactions. Please note that since these interactions are unpaired, the direction of the linkers has no real spatial meaning. Ligand linkers are directed away from the ligand, while protein linkers are directed towards the ligand.
Table 5. Examples of visualizing unpaired salt-bridge interactions 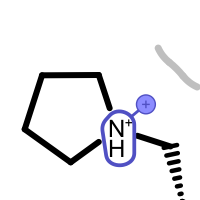
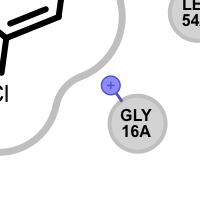
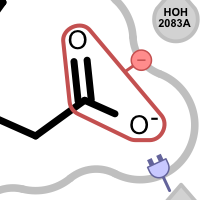
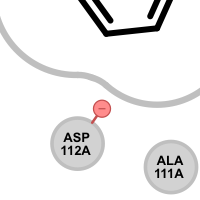
unpaired ligand positive
unpaired protein positive
unpaired ligand negative
unpaired protein negative
See also
OESaltBridgeInteractionHint class in the OEChem TK manual
OESaltBridgeInteractionHintType namespace in the OEChem TK manual
Hydrogen Position Optimization
Since the interaction perception dependents on the position of hydrogens, it is highly recommended to optimize those positions prior to perceiving the interactions. The two images below reveal the effect of optimizing the hydrogen bond network in a protein-ligand complex: less atom clash(es) and less unpaired hydrogen bond interaction(s).
original complex downloaded from PDB Database (PDB 1D3H) |
complex after optimizing hydrogen positions |
See also
OEPlaceHydrogens function in the OEChem TK manual
Protein Preparation chapter in the OEChem TK manual
Unpaired Map vs Active Site Interaction Map
An unpaired interaction map provides a complementary view to the more common active site interaction map. While the interaction map (on the right) depicts interactions between the ligand and protein, the unpaired map (on the left) illustrates interactions that could contribute to binding but are not formed in the complex. Together, these two maps of the protein-ligand binding site provide insights into protein-ligand interactions and communicate complex 3D structural results to medicinal chemists in a directly actionable way.
unpaired interaction map (PDB 1BR6) |
active site interaction map (PDB 1BR6) |
See also in OEChem TK manual
Theory
Biopolymers chapter
Protein Preparation chapter
API
OEPerceiveInteractionHints function
OEPlaceHydrogens function
See also in OEDepict TK manual
Theory
Molecule Depiction chapter
API
OEImage class
See also in GraphemeTM TK manual
API
OE2DActiveSiteDisplay class
OEDrawUnpairedInteractionMapLegend function
OEPrepareActiveSiteDepiction function
OERenderUnpairedInteractionMap function