Rapid Similarity Searching of Large Molecule Files¶
Problem¶
You want to perform rapid 2D similarity search on large molecule files.
Ingredients¶
|
Download¶
Download code
makefastfp.py to generate binary fast fingerprint files
searchfastfp.py to search binary fast fingerprint files
See also the Usage (makefastfp) and Usage (searchfastfp) subsections.
Solution¶
In order to solve this problem two databases are utilized:
- The OEMolDatabase class of OEChem TK is used is to provide fast read-only random access to molecular file formats that are readable by OEChem TK. This class is designed to handle large molecule files that can not be held in memory all at once.
- The OEFastFPDatabase class of GraphSim TK is used to perform rapid in-memory or memory-mapped fingerprint search using popcount method.
See also
- Manipulating Large Molecule Files recipe for the introduction to the OEMolDatabase class.
This recipe discusses two code examples:
- Generating Fingerprints section shows how to generate fingerprints and save them into a binary fingerprint file.
- Searching Fingerprints section illustrates how to performed rapid similarity search using the pre-generated fingerprints.
Generating Fingerprints¶
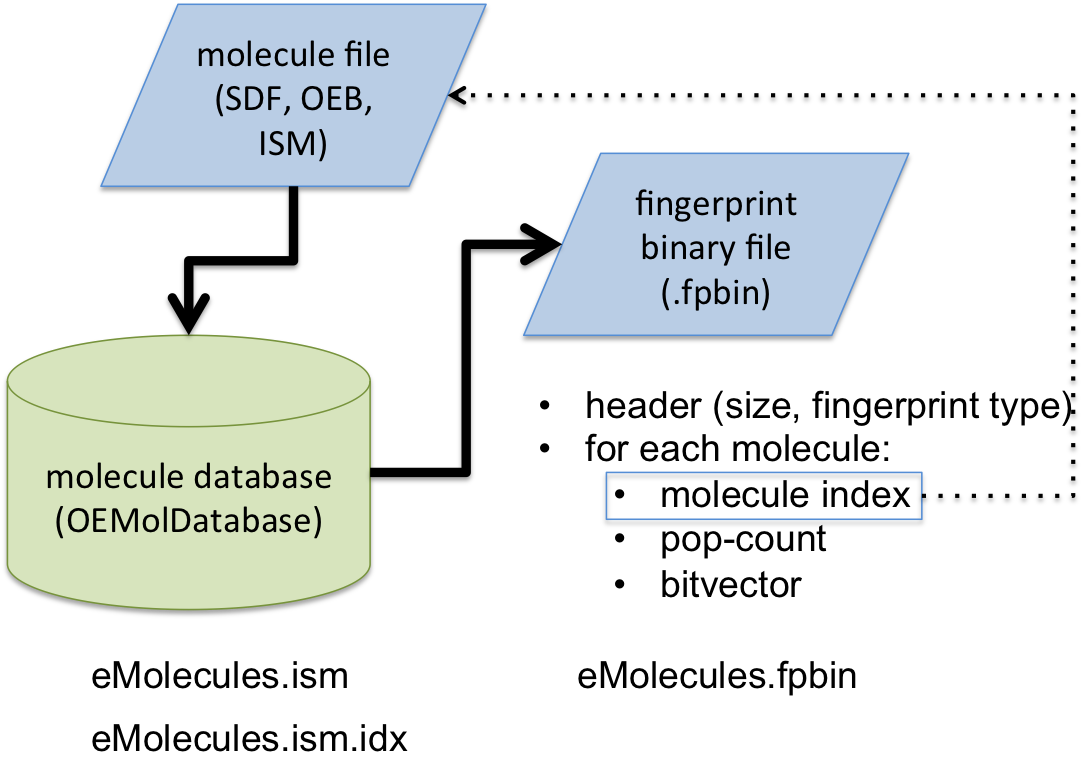
In order to accelerate the molecule similarity search, fingerprints that encode local features of molecules are generated and stored in a binary file with a .fpbin extension. The following code snippet shows how to generate a binary fingerprint file. See Figure 1. that illustrates the overall process of the fingerprint generation.
First, an OEMolDatabase object is initialized with a input molecule filename (lines 1-3), followed by the initialization of fingerprint type from the command line interface (lines 5-6) using the OESetupFingerPrint function. A binary fingerprint file is then generated by the OECreateFastFPDatabaseFile function (line 16). The fingerprint generation is multi-thread. The number of processors that are utilized can be controlled via the OECreateFastFPDatabaseOptions class. (See also: Performance of fingerprint generation section)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | ifname = itf.GetString("-in")
ffname = itf.GetString("-fpdb")
if oechem.OEGetFileExtension(ffname) != "fpbin":
oechem.OEThrow.Fatal("Fingerprint database file should have '.fpbin' file extension!")
idxfname = oechem.OEGetMolDatabaseIdxFileName(ifname)
if not os.path.exists(idxfname):
if not oechem.OECreateMolDatabaseIdx(ifname):
oechem.OEThrow.Warning("Unable to create %s molecule index file", idxfname)
print("Using %s index molecule file" % idxfname)
moldb = oechem.OEMolDatabase()
if not moldb.Open(ifname):
oechem.OEThrow.Fatal("Cannot open molecule database file!")
fptype = oegraphsim.OESetupFingerPrint(itf)
|
The generated binary fingerprint file (with .fpbin extension) stores the following information for each molecule:
- the index of the corresponding molecule in the OEMolDatabase object
- the bitvector of the fingerprint
- the popcount of the fingerprint i.e the number of bits that are set in the given fingerprint
Usage (makefastfp)¶
The first example shows how to generate a binary fingerprint file that stores tree fingerprints (that is the default type) for the eMolecules database:
- Retrieve eMolecules dataset in SMILES format from web server
- Unzip file, remove first line (header) and only keep the SMILES without title
- Generate tree fingerprints using makefastfp.py
Usage
1 2 3 | prompt > wget http://downloads.emolecules.com/free/2018-02-01/version.smi.gz
prompt > gzip -dc version.smi.gz | awk '{if (NR!=1) {print $1 }}' > eMolecules.ism
prompt > python3 makefastfp.py -in eMolecules.ism -fpdbfname eMolecules-tree.fpbin
|
Running the above commands will generate the following output:
Using eMolecules.ism.idx index molecule file
Using fingerprint type Tree,ver=2.0.0,size=4096,bonds=0-4,atype=AtmNum|Arom|Chiral|FCharge|HvyDeg|Hyb,btype=Order
Generating fingerprints with 16 threads
.....
Total: 10697014 fingerprints processed.
268.83 secs to generate 10697014 fingerprints
The generated eMolecules-tree.fpbin file stores not only fingerprints but also information about:
- the endianess of the binary fingerprint file
- the name of the molecule file to which the fingerprint file corresponds
- the string representation of the fingerprint type
- the number of fingerprints in the binary file
This information can be accessed via the OEFastFPDatabaseParams class or by peeking into the fingerprint file:
prompt > head -4 eMolecules-tree.fpbin
LE
eMolecules.ism
Tree,ver=2.0.0,size=4096,bonds=0-4,atype=AtmNum|Arom|Chiral|FCharge|HvyDeg|Hyb,btype=Order
10697014
Note
The fast fingerprint search does not support endianness compatibility for performance reason. That means that a binary fingerprint file that has been generated in a little-endian computer cannot be search in a computer that uses the big-endian encoding.
The fast fingerprint search also supports circular or path fingerprint types. The following examples show how to generate the corresponding binarry fingerprint files:
Usage
> python3 makefastfp.py -in eMolecules.ism -fpdbfname eMolecules-path.fpbin -fptype Path
> python3 makefastfp.py -in eMolecules.ism -fpdbfname eMolecules-circ.fpbin -fptype Circular
Note
GraphSim TK currently only supports popcount search method for fingerprints with the size of multiple of 256. This means that the MACCS key fingerprint type is currently not supported. When customizing other fingerprint types such as tree, circular path , the size of the fingerprint must be a multiple of 256.
See also User-defined Fingerprint section in the GraphSim TK manual.
The following example shows how to generate custom fingerprints by specifying:
- the size of the fingerprint (using -numbits parameter)
- the atom and bond properties that are encoded into each fingerprint (using -atomtype and -bondtype parameters, respectively)
Usage
> python3 makefastfp.py -in eMolecules.ism -fpdbfname eMolecules-custom.fpbin -fptype Tree -numbits 1024 -atomtype "AtmNum|Arom|HvyDeg|" -bondtype "Order|InRing"
Using eMolecules.ism.idx index molecule file
Using fingerprint type Tree,ver=2.0.0,size=1024,bonds=0-4,atype=AtmNum|Arom|HvyDeg,btype=Order|InRing
Generating fingerprints with 16 threads
.....
See also
Searching Fingerprints¶
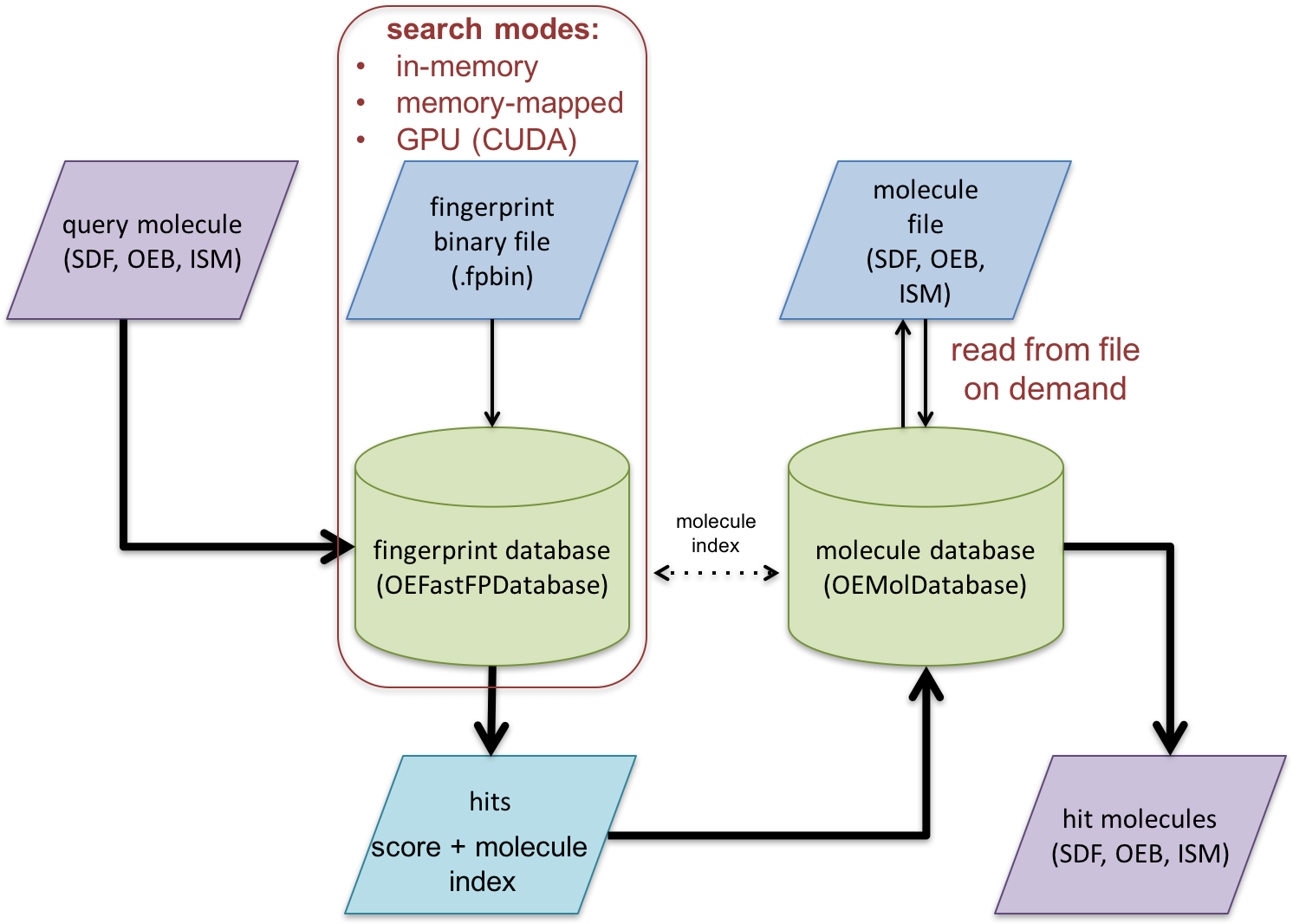
The following code example illustrates how to search the binary fingerprint file generated by the previous example. See Figure 2. that illustrates the overall fingerprint search process.
In this example two databases are utilized:
- An OEMolDatabase object is initialized from a molecule file (lines 1-3).
- An OEFastFPDatabase object is initialized from a binary fingerprint file (lines 5-14). Fingerprint databases can be initialized in either in-memory or memory-mapped mode. For more information see the Memory-Mapped vs In-Memory Search section.
The correspondence between the two databases are maintained by molecule indexes (see Figure 2.). This correspondence can be checked by calling the OEAreCompatibleDatabases function (lines 16-17).
After initializing the databases, OEFPDatabaseOptions object is created (lines 24-25) that controls how the fingerprint database is searched (lines 29-31). For more details about the search options see the Fingerprint search options section The OEFastFPDatabase.GetSortedScores method returns an iterator over the calculated similarity scores (OESimScore) in sorted order. Each OESimScore holds a similarity score and index of the corresponding fingerprint of the database. This index can be used to access the original molecule in the OEMolDatabase object (lines 33-39) and write the hits into an output molecule file.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 | # initialize databases
timer = oechem.OEWallTimer()
timer.Start()
ifs = oechem.oemolistream()
if not ifs.open(qfname):
oechem.OEThrow.Fatal("Cannot open input file!")
ofs = oechem.oemolostream()
if not ofs.open(ofname):
oechem.OEThrow.Fatal("Cannot open output file!")
query = oechem.OEGraphMol()
if not oechem.OEReadMolecule(ifs, query):
oechem.OEThrow.Fatal("Cannot read query molecule!")
moldb = oechem.OEMolDatabase()
if not moldb.Open(mfname):
oechem.OEThrow.Fatal("Cannot open molecule database!")
memtype = oegraphsim.OEGetFPDatabaseMemoryType(itf)
fpdb = oegraphsim.OEFastFPDatabase(ffname, memtype)
if not fpdb.IsValid():
oechem.OEThrow.Fatal("Cannot open fingerprint database!")
nrfps = fpdb.NumFingerPrints()
memtypestr = fpdb.GetMemoryTypeString()
if not oegraphsim.OEAreCompatibleDatabases(moldb, fpdb):
oechem.OEThrow.Fatal("Databases are not compatible!")
print("%5.2f sec to initialize databases" % timer.Elapsed())
fptype = fpdb.GetFPTypeBase()
print("Using fingerprint type %s" % fptype.GetFPTypeString())
opts = oegraphsim.OEFPDatabaseOptions()
oegraphsim.OESetupFPDatabaseOptions(opts, itf)
|
Usage (searchfastfp)¶
The following example shows how to search molecules that are similar to caffeine in the eMolecules database using the memory-mapped mode. The SMILES string of the query molecule is read from the standard input (-query .ism) and the SMILES of the top ten most similar molecules are written into standard output (-out .ism).
Usage
prompt > python3 searchfastfp.py -fpdb eMolecules-tree.fpbin -molfname eMolecules.ism -query .ism -out .ism
c12c(n(c(n(c1=O)C)=O)C)ncn2C
Running the above command will generate the following output:
4.75 sec to initialize databases
Using fingerprint type Tree,ver=2.0.0,size=4096,bonds=0-4,atype=AtmNum|Arom|Chiral|FCharge|HvyDeg|Hyb,btype=Order
5.07 sec to search 10697015 fingerprints memory-mapped
Cn1cnc2c1c(=O)n(c(=O)n2C)N
Cn1c2c(c(=O)n(c1=O)C)n(cn2)N
Cn1cnc2c1c(=O)n(c(=O)n2C)Cn3c(=O)c4c(ncn4C)n(c3=O)C
Cn1cnc2c1c(=O)n(c(=O)n2C)CCn3c(=O)c4c(ncn4C)n(c3=O)C
Cn1cnc2c1c(=O)n(c(=O)n2C)CC#C
Cn1cnc2c1c(=O)n(c(=O)n2C)CCCO
Cn1cnc2c1c(=O)n(c(=O)n2C)CC=C
CCn1cnc2c1c(=O)n(c(=O)n2C)C
Cn1c2c(c(=O)n(c1=O)C)n(cn2)C=C
Cn1cnc2c1c(=O)n(c(=O)n2C)CCCBr
When the hits are saved into an sdf file, the similarity scores will be written to each hit with the <Similarity score> tag. The in-memory mode can be set using the -memorytype parameter.
Usage
prompt > python3 searchfastfp.py -fpdb eMolecules-tree.fpbin -molfname eMolecules.ism -query caffeine.ism -out hits.sdf -memorytype in-memory
See also
Discussion¶
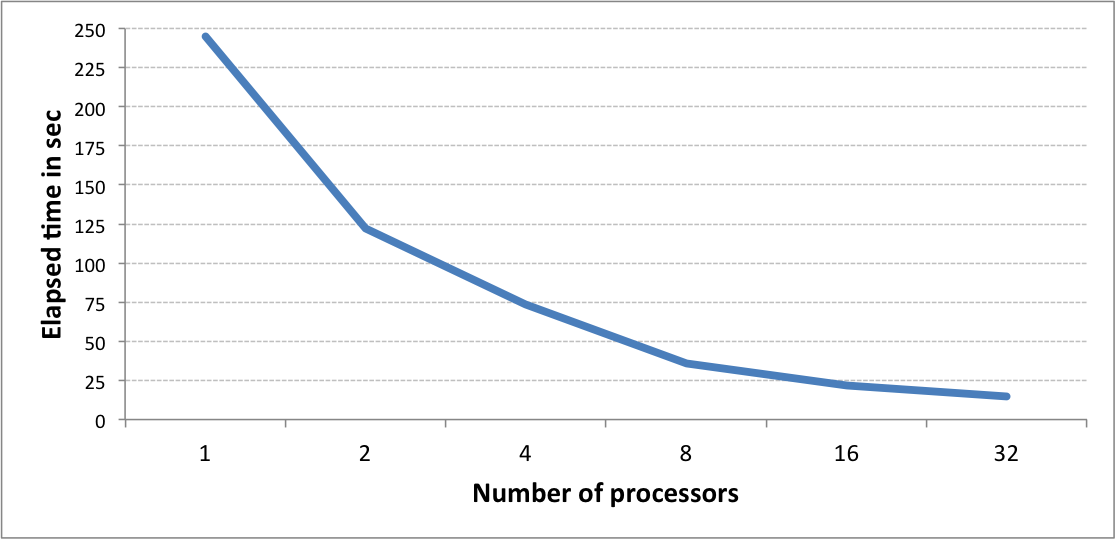
Performance of fingerprint generation¶
The graph below shows the speed of generating 1 million 4096-bit long tree fingerprints using different number of processes. The performance has been benchmarked using m4.10x large AWS instance.
Fingerprint search options¶
The OEFastFPDatabase class provides several ways to search fingerprints by utilizing the OEFPDatabaseOptions class:
By default, OETanimoto similarity coefficient is used to quantify the degree of resemblance between two fingerprints. However, other built-in similarity coefficients such as OECosine, OEDice, OEEuclidean, OEManhattan, and OETversky can also be used (see also OEFPDatabaseOptions.SetSimFunc method).
Note
The current implementation of fast fingerprint search method does not allow to utilize user-defined similarity measure. If this is desired the slower fingerprint search implemented in the OEFPDatabase class should be used.
By default, the OEFPDatabase.GetSortedScores method returns similarity scores in descending order i.e. it identifies molecules that are most similar to the query. However, the order can be reversed to identify molecules that are most dissimilar to the query (see also OEFPDatabaseOptions.SetDescendingOrder method).
There is no default cut-off value defined for fingerprint search, since a reasonable cut-off value can depend on various parameters such as: the fingerprint type, the query molecule, and the the similarity measure used. The user can specify a cut-off value for a specific search using the OEFPDatabaseOptions.SetCutOff method.
By default, there is no limit for number of similarity scores returned, however searching and ordering millions of molecules are not recommended. A reasonable limit can be set by using the OEFPDatabaseOptions.SetLimit method.
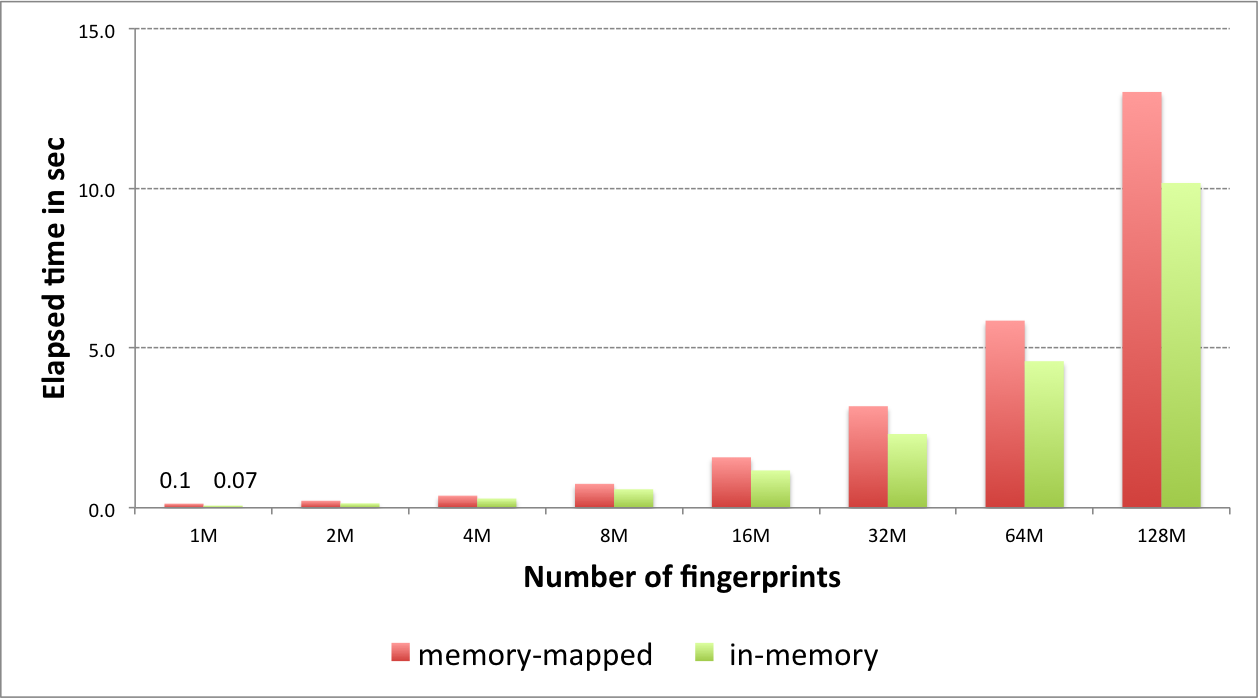
Performance of fingerprint search¶
The graph below shows the speed of searching 4096-bit long tree fingerprints for increasing fingerprint database size.
|
|
Note
The fast fingerprint search implemented by the OEFastFPDatabase class has been optimized for OETanimoto that is the most widely used similarity measure.
Memory-Mapped vs In-Memory Search¶
The OEFastFPDatabase class can be initialized in two modes:
- in-memory
- The in-memory mode involves pre-loading all fingerprints into memory prior to and performing the search in the memory. While this represents the fastest way to perform similarity searches once the fingerprints are loaded, searches are limited by memory availability.
- memory-mapped
- The memory-mapped mode has no load time penalty or memory limitation but the search itself takes more time.
In order to demonstrate the performance difference between the in-memory and the memory-mapped modes, the same search using the is carried out on m4.10x large AWS instance:
Output of the search when using the in-memory mode:
4.70 sec to initialize databases
Using fingerprint type Tree,ver=2.0.0,size=4096,bonds=0-4,atype=AtmNum|Arom|Chiral|FCharge|HvyDeg|Hyb,btype=Order
0.73 sec to search 10697015 fingerprints in-memory
Output of the search with using the memory-mapped mode:
0.15 sec to initialize databases
Using fingerprint type Tree,ver=2.0.0,size=4096,bonds=0-4,atype=AtmNum|Arom|Chiral|FCharge|HvyDeg|Hyb,btype=Order
0.93 sec to search 10697015 fingerprints memory-mapped
The in-memory search can be %10-%25 faster then the memory-mapped search, but the startup ‘cost’ of the in-memory search is greater.
See also in OEChem TK manual¶
Theory
- Molecular Database Handling chapter
API
- OECreateMolDatabaseIdx function
- OEMolDatabase class
See also in GraphSim TK manual¶
Theory
- Fingerprint Generation chapter
API
- OEAreCompatibleDatabases function
- OEConfigureFingerPrint and OESetupFingerPrint functions
- OEConfigureFPDatabaseOptions and OESetupFPDatabaseOptions functions
- OECreateFastFPDatabaseFile function
- OECreateFastFPDatabaseOptions class
- OEFastFPDatabase class
- OEFastFPDatabaseParams class
- OEFingerPrint class
- OEFPDatabaseOptions class
- OESimScore class