Fingerprint Generation
The fingerprint types implemented in the GraphSim TK encode the 2D graph features of molecules. Fingerprints can be used in applications such as similarity searches, molecular characterization, molecular diversity and chemical database clustering.
The following five types of fingerprints are implemented:
MACCS
MACCS keys are 166 bit structural key descriptors in which each bit is associated with a SMARTS pattern.
The following code snippets demonstrate two separate ways to create a MACCS keys fingerprint:
fp = oegraphsim.OEFingerPrint()
oegraphsim.OEMakeMACCS166FP(fp, mol)
oegraphsim.OEMakeFP(fp, mol, oegraphsim.OEFPType_MACCS166)
LINGO
The GraphSim TK provides fingerprint API for the LINGO similarity search method implemented in OEChem TK.
The following code snippets demonstrate two separate ways to create a LINGO fingerprint:
fp = oegraphsim.OEFingerPrint()
oegraphsim.OEMakeLingoFP(fp, mol)
oegraphsim.OEMakeFP(fp, mol, oegraphsim.OEFPType_Lingo)
Circular
A circular fingerprint is generated by exhaustively enumerating all circular fragments grown radially from each heavy atom of the molecule up to the given radius and then hashing these fragments into a fixed-length bitvector. See Figure: Example of enumerating circular fragments with various radii.
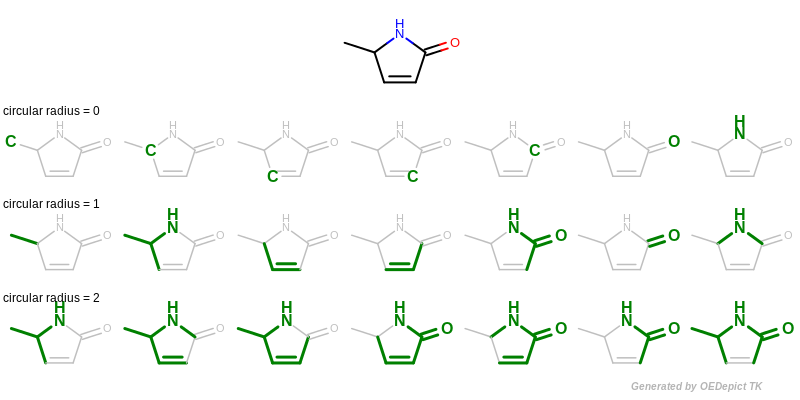
Example of enumerating circular fragments with various radii
The following code snippets demonstrate two separate ways to create a circular fingerprint with default parameters:
fp = oegraphsim.OEFingerPrint()
oegraphsim.OEMakeCircularFP(fp, mol)
oegraphsim.OEMakeFP(fp, mol, oegraphsim.OEFPType_Circular)
Path
A path fingerprint is generated by exhaustively enumerating all linear fragments of a molecular graph up to a given size and then hashing these fragments into a fixed-length bitvector. See Figure: Example of enumerating path fragments with various lengths.
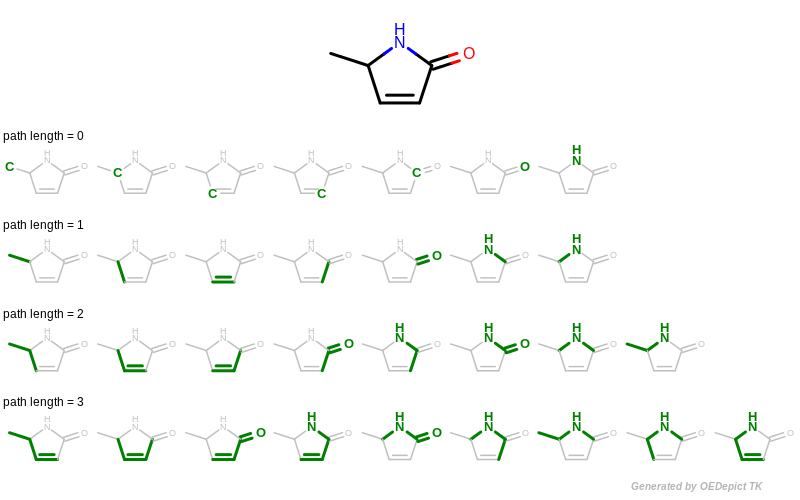
Example of enumerating path fragments with various lengths
The following code snippets demonstrate two separate ways to create a path fingerprint with default parameters:
fp = oegraphsim.OEFingerPrint()
oegraphsim.OEMakePathFP(fp, mol)
oegraphsim.OEMakeFP(fp, mol, oegraphsim.OEFPType_Path)
Tree
A tree fingerprint is generated by exhaustively enumerating all tree fragments of a molecular graph up to a given size and then hashing these fragments into a fixed-length bitvector. See Figure: Example of enumerating tree fragments with various lengths.
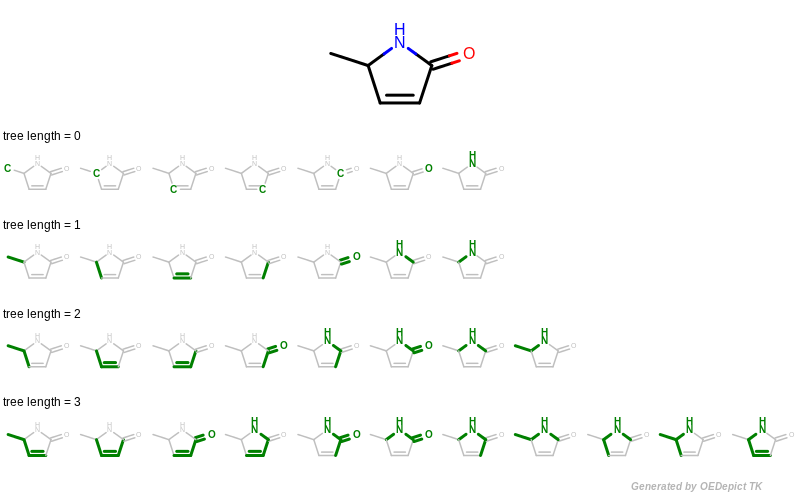
Example of enumerating tree fragments with various lengths
The following code snippets demonstrate two separate ways to create a tree fingerprint with default parameters:
fp = oegraphsim.OEFingerPrint()
oegraphsim.OEMakeTreeFP(fp, mol)
oegraphsim.OEMakeFP(fp, mol, oegraphsim.OEFPType_Tree)
Hint
GraphSim TK also provides the ability to parameterize the circular, path and tree fingerprint generation with arbitrary sets of properties. For more details see the User-defined Fingerprint chapter.