OEFastFPDatabase
class OEFastFPDatabase
The OEFastFPDatabase class is designed to perform rapid CUDA-accelerated, in-memory or memory-mapped fingerprint searches using the popcount method. Each OEFastFPDatabase object is associated with a fingerprint type (OEFPTypeBase) that is set when the database is initialized from a pre-generated binary fingerprint file.
For CUDA-accelerated fingerprint searching please see the prerequisites for OpenEye’s GPU-accelerated software.
Note
GraphSim TK currently only supports the popcount search
method for fingerprints with the size of multiple of 256.
This means that the OEFastFPDatabase class
currently does not support:
OEFPType_MACCS166fingerprint typeOEFPType_Lingofingerprint typeother custom fingerprint types (see User-defined Fingerprint section) with size that is not a multiple of
256
For the fingerprint types listed above, the original OEFPDatabase class can be utilized.
Note
OEFastFPDatabase gives identical results to OEFPDatabase. However, OEFPDatabase calculates similarity scores in single precision (float) while OEFastFPDatabase uses double precision. As a result, small similarity score differences can be observed.
See also
OEFPDatabase class
Code Example
Example code in the Generating fingerprint file for fast fingerprint search section
Example code in the Searching fast fingerprint database section
Rapid Similarity Searching of Large Molecule File OpenEye Python Cookbook recipe
Depicting Molecule Similarity Based on Fingerprints OpenEye Python Cookbook recipe
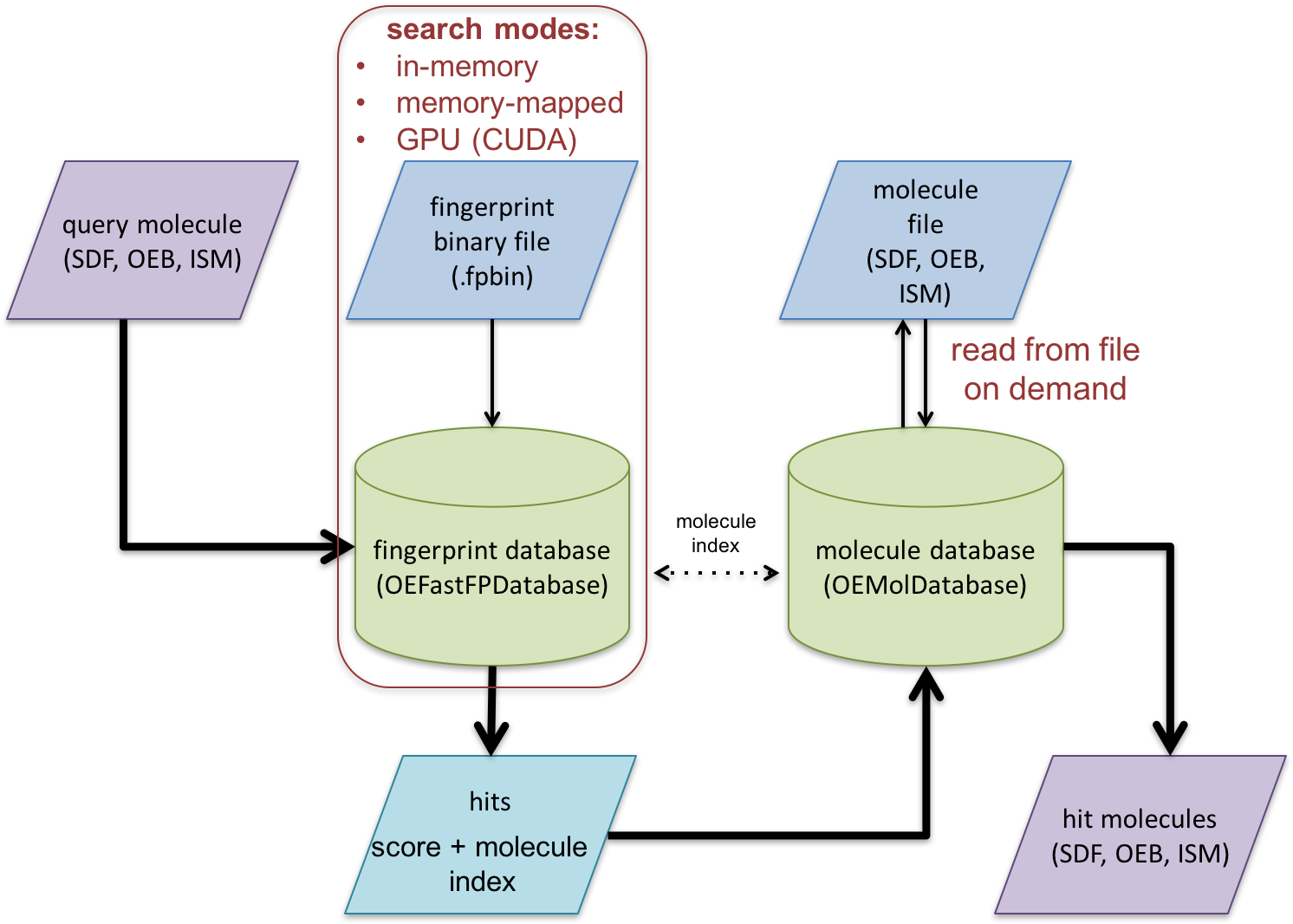
Schematic representation of fast fingerprint search process
Constructors
OEFastFPDatabase(const std::string &dbfile,
unsigned int memtype=OEFastFPDatabaseMemoryType::Default)
Constructs an OEFastFPDatabase object.
- dbfile
The name of the file which contains the fingerprint data. The file has to be generated with the
OECreateFastFPDatabaseFilefunction.- memtype
Defines whether the fingerprints are pre-loaded into GPU-memory, CPU-memory, or memory-mapped during the search process. This value has to be from the
OEFastFPDatabaseMemoryTypenamespace.
Note
If the OEFastFPDatabase object can not be initialized
with the OEFastFPDatabaseMemoryType_CUDA option,
the following warning message will be throw:
Warning: OEFastFPDatabase::OEFastFPDatabase() : no CUDA-enabled
device available falling back to memory-mapped type!
As a rule of thumb, 1 million finger prints requires 0.6GB of GPU memory.
GPU memory can be queried using the nvidia-smi command from terminal.
See also
OEFastFPDatabaseMemoryTypenamespace.
GetAllScores
OESystem::OEIterBase<float> *GetAllScores(const OEFPDatabaseOptions &opts) const
Performs \(NxN\) similarity calculations between all pairs of fingerprints stored in the OEFastFPDatabase object. It returns an iterator over the calculated similarity scores. The scores are not sorted, but returned as a flattened square matrix.
For all similarity measures other than Tversky, the returned ‘matrix’ will be symmetrical.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the search (i.e. similarity measure parameters). Cutoff and order parameters are ignored as the results are not filtered or sorted.
Note
This operation scales with \(n^2\) in memory and can easily overwhelm
the system memory for larger databases.
Therefore, the OEFastFPDatabase.GetRawScores method
returns a single row of the similarity matrix and is the recommended usage
for querying large databases.
See also
OEFPDatabaseOptions class
GetFingerPrint
bool GetFingerPrint(OEFingerPrint& fp, size_t idx) const
Returns the \(idx^{th}\) fingerprint of the database.
Warning
This function returns false if the fingerprint index is
not identical to the corresponding molecule index.
This can occur if the fingerprint binary file is generated in
multi-thread process.
If direct access to the fingerprint is required when using
the OEFastFPDatabase.GetFingerPrint
method, the fingerprint file should be generated in a
single-threaded mode.
This can be done by setting
SetNumProcessors(1)
for the option class used for creating the binary file.
See also
GetFPTypeBase
const OEFPTypeBase *GetFPTypeBase() const
Returns the fingerprint type of the OEFastFPDatabase object. An OEFastFPDatabase object can only store fingerprints with identical types.
GetHistogram
OEFPHistogram *GetHistogram(const OEFPDatabaseOptions &opts,
const size_t nrbins=200u) const
Performs similarity calculations between all pairs of fingerprints stored in the OEFastFPDatabase object. It returns the histogram over the calculated similarity scores in a OEFPHistogram object.
- opts
The OEFPDatabaseOptions object contains the settings available to control the search (i.e. similarity measure, \(\alpha\) and \(\beta\) parameters for Tversky similarity). Cutoff and order parameters are ignored as the results are not filtered or sorted.
- nrbins
Number of bins in the returned OEFPHistogram object.
Note
For all similarity measures other than Tversky, the histogram only contains the upper-triangular similarity scores (excluding the diagonal). In case of the asymmetric Tversky similarity measure, the histogram of the whole \(NxN\) matrix is returned.
Note
When the OEFastFPDatabase object is initialized
with OEFastFPDatabaseMemoryType_CUDA,
nrbins is limited to at most 1024.
Hint
This method calculates similarities identically to
OEFastFPDatabase.GetAllScores
but is not bound by the system memory. It can be used to quickly obtain
statistics on larger databases.
See also
OEFPDatabaseOptions class
OEFPHistogram class
GetMemoryType
unsigned int GetMemoryType() const
Returns the memory type of the fingerprint database.
The return value is taken from the OEFastFPDatabaseMemoryType
namespace.
See also
GetMemoryTypeString
std::string GetMemoryTypeString() const
Returns the string representation if memory type of the fingerprint database.
See also
GetMoleculeIndex
size_t GetMoleculeIndex(const size_t fpidx) const
Returns the molecule index that corresponds to the fingerprint index.
Note
When building fingerprint databases using
OECreateFastFPDatabaseOptions, the molecule
index is always the same as the fingerprint index. However, there
are private database building APIs that allow to specify the molecule
index associated with each fingerprint.
This OEFastFPDatabase.GetMoleculeIndex
method allows to handle these private databases.
GetRawScores
OESystem::OEIterBase<double> *GetRawScores(const size_t fpidx,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<double> *GetRawScores(const OEFingerPrint &fp,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<double> *GetRawScores(const OEChem::OEMolBase &mol,
const OEFPDatabaseOptions &opts) const
Performs similarity calculations between a molecule or a fingerprint and the fingerprints stored in the OEFastFPDatabase object. It returns an iterator over the calculated similarity scores. The scores are not sorted, but returned in the same order as the database. The number of elements in the returned iterator is equal to the number of fingerprints in the database.
- fpidx
If the method is called with an integer index, the query fingerprint is taken from the OEFastFPDatabase object with the given index.
- mol
If the method is called with an OEMolBase object, a fingerprint is generated from this molecule before looping over the fingerprints of the database and calculating similarities.
- fp
If the method is called with an OEFingerPrint object, its type has to match the type of the OEFastFPDatabase.
- opts
The OEFPDatabaseOptions object contains the settings available to control the search (i.e. similarity measure parameters). Cutoff and order parameters are ignored as the results are not filtered or sorted.
See also
OEFPDatabaseOptions class
GetScores
OESystem::OEIterBase<OESimScore> *GetScores(const size_t idx,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<OESimScore> *GetScores(const OEFingerPrint &fp,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<OESimScore> *GetScores(const OEChem::OEMolBase &mol,
const OEFPDatabaseOptions &opts) const
Performs similarity calculations between a molecule or fingerprint and the
fingerprints stored in the OEFastFPDatabase object.
It returns an iterator over the calculated similarity scores
(OESimScore).
The results are filtered according to the cutoff and order parameters
specified in opts, but are not sorted.
- fpidx
If the method is called with an integer index, the query fingerprint is taken from the OEFastFPDatabase object with the given index.
- mol
If the method is called with an OEMolBase object, a fingerprint is generated from this molecule before looping over the fingerprints of the database and calculating similarities.
- fp
If the method is called with an OEFingerPrint object, its type has to match the type of the OEFastFPDatabase.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the search (i.e. similarity measure parameters, score cutoff and order).
See also
OEFPDatabaseOptions class
GetSortedScores
OESystem::OEIterBase<OESimScore> *GetSortedScores(const size_t idx,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<OESimScore> *GetSortedScores(const OEFingerPrint &fp,
const OEFPDatabaseOptions &opts) const
OESystem::OEIterBase<OESimScore> *GetSortedScores(const OEChem::OEMolBase &mol,
const OEFPDatabaseOptions &opts) const
Performs similarity calculations between a molecule or fingerprint and the fingerprints stored in the OEFastFPDatabase object. It returns an iterator over the calculated similarity scores (OESimScore) in sorted order. Each OESimScore holds a similarity score and index of the corresponding fingerprint of the database.
- fpidx
If the method is called with an integer index, the query fingerprint is taken from the OEFastFPDatabase object with the given index.
- mol
If the method is called with an OEMolBase object, a fingerprint is generated from this molecule before looping over the fingerprints of the database and calculating similarities.
- fp
If the method is called with an OEFingerPrint object, its type has to match the type of the OEFastFPDatabase.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the search (i.e. similarity measure parameters, score cutoff, order and limit).
See also
OEFPDatabaseOptions class
GetSparseMatrix
OESystem::OEIterBase<OESimScorePair> *GetSparseMatrix(const OEFPDatabaseOptions &opts) const
Performs \(NxN\) similarity calculations between all pairs of fingerprints
stored in the OEFastFPDatabase object and returns either the top K
scores for each fingerprint or all scores above a cutoff for each fingerprint. The sparse
matrix is returned as an iterator over OESimScorePair objects. The
limit of scores to return can be set using OEFPDatabaseOptions.SetLimit
and the cutoff of scores to return can be set using OEFPDatabaseOptions.SetCutoff.
If no limit is set, all scores will be returned above the cutoff value. If a limit is set, only
OEFPDatabaseOptions.GetLimit scores will be returned regardless of whether
more scores fall within the cutoff range. A limit should be set for best performance.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the search (i.e. similarity measure parameters).
Hint
This operation scales with \(n^2\) in memory when
limit = 0 (or when limit is not set), so it can easily overwhelm
the system memory for larger databases. The best practice is to set a
reasonable limit that will capture the scores of interest.
See also
OEFPDatabaseOptions class
OESimScorePair class
GetVariogram
OEGraphSim::OEFPVariogram *GetVariogram(const std::vector<float>& obsdata,
const OEFPDatabaseOptions &opts,
const size_t nrbins=200u) const
Performs similarity calculations between all pairs of fingerprints
stored in the OEFastFPDatabase object.
It returns the empirical variogram over the calculated similarity scores
with respect to the measurements provided in the obsdata parameter in a
OEFPVariogram object.
- obsdata
User-provided empirical measurements for each fingerprint in the database.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the scoring (i.e. similarity measure parameters). Cutoff and order parameters are ignored as the results are not filtered or sorted.
- nrbins
Number of bins in the returned OEFPVariogram object.
Note
The empirical variogram is defined over distances rather than similarities. It is therefore not possible to calculate a variogram using Tversky similarity. For all other similarity measures, empirical variogram is calculated using \(distance = 1-similarity\).
Hint
This method calculates similarities identically to
OEFastFPDatabase.GetAllScores
but is not bound by the system memory. It can be used to quickly obtain
statistics on larger databases.
Note
When the OEFastFPDatabase object is initialized
with OEFastFPDatabaseMemoryType_CUDA,
nrbins is limited to at most 1024.
Hint
The returned OEFPVariogram object also contains a histogram, but note that this histogram is over distances rather than similarities.
See also
OEFPDatabaseOptions class
OEFPVariogram class
IsValid
bool IsValid() const
Returns whether the database was initialized correctly.
NumFingerPrints
size_t NumFingerPrints() const
Returns the number of OEFingerPrint objects stored in the database.
SortedSearch
unsigned SortedSearch(OESimSearchResult &result,
const OEChem::OEMolBase &mol,
const OEFPDatabaseOptions &opts) const
Performs multi-threaded similarity calculations between a molecule and the
fingerprints stored in the OEFastFPDatabase object.
The method combines the functionality of the
OEFastFPDatabase.GetSortedScores and the
OEFastFPDatabase.GetHistogram methods.
- result
The OESimSearchResult object that stores the result of the search along with the progress of the search and the histogram of all scores.
- mol
If the method is called with an OEMolBase object, a fingerprint is generated from this molecule before looping over the fingerprints of the database and calculating similarities.
- opts
The OEFPDatabaseOptions object controls all the parameters that determine the search (i.e. similarity measure parameters). The
OEFastFPDatabase.SortedSearchmethod can use multiple threads to accelerate the search process. The number of processors used can be controlled by theOEFPDatabaseOptions.SetNumProcessorsmethod.
The OEFastFPDatabase.SortedSearch method returns:
OESimSearchStatus_Uninitializedif the similarity search can not be executedOESimSearchStatus_Finished, if the similarity search successfully finished
Note
This method is currently only available in
OEFastFPDatabaseMemoryType_MemoryMapped and
OEFastFPDatabaseMemoryType_InMemory modes.
Example:
query = oechem.OEGraphMol()
oechem.OESmilesToMol(query, "Cc1c(c2cc(ccc2n1C(=O)c3ccc(cc3)Cl)OC)CC(=O)O")
limit = 5
opts = oegraphsim.OEFPDatabaseOptions(limit, oegraphsim.OESimMeasure_Tanimoto)
nrbins = 5
result = oegraphsim.OESimSearchResult(nrbins)
status = fpdb.SortedSearch(result, query, opts)
print("Search status = {}".format(oegraphsim.OESimSearchStatusToName(status)))
print("Number of searched = {}".format(result.NumSearched()))
# print scores
for score in result.GetSortedScores():
print("{:.3f}".format(score.GetScore()))
# print histogram
hist = result.GetHistogram()
bounds = hist.GetBinBoundaries()
for idx, count in enumerate(hist.GetCounts()):
print("[{:.3f}-{:.3f}] = {}".format(bounds[idx], bounds[idx+1], count))
The output of the code snippet above might look like this:
Search status = Finished
Number of searched = 1000
0.950
0.912
0.901
0.886
0.859
[0.000-0.200] = 428
[0.200-0.400] = 312
[0.400-0.600] = 225
[0.600-0.800] = 25
[0.800-1.000] = 10
See also
OESimSearchStatusnamespace