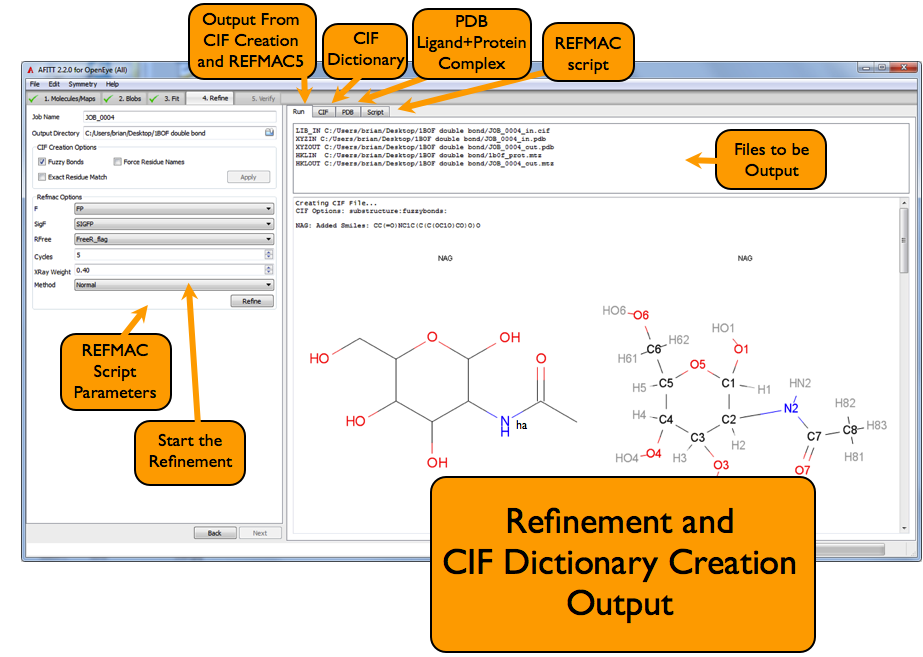
Refine Complex: Refine Page¶
AFITT is tightly integrated to work with either REFMAC or PHENIX as refinement programs. If the refinement program is properly installed and the refinement workflow is set up, AFITT can launch it and validate its output. If the refinement program is not installed, AFITT will save the protein complex, refinement dictionary and script to the desired location, but no refinement will be done. These modes are called, automatic and manual. To recap:
- AUTOMATIC mode: allows AFITT to run refmac or phenix.refine and validate the results.
- GENERATE Refinement Scripts mode: saves the pdb and dictionary to run outside of AFITT.
Both the automatic and manual modes look similar, the only difference is that in automatic mode there is a Refine button as part of the refinement parameters and for the manual mode, this is replaced with a Save button.
Refinement Dictionary¶
In either mode, AFITT always generates a refinement dictionary automatically. The refinement dictionary report is shown in the Run tab of the widget as shown in Figure REFMAC5 Refinement Page or Figure PHENIX Refinement Page.
This report describes the residue dictionary that were created, and whether they mapped to a known residue or had to be replaced with a new dictionary and residue name.
The refinement dictionary itself, is viewable in the CIF tab while the PDB structure is viewable in the PDB tab.
Refinement Parameters¶
Refinement parameters can be modified to alter the generated script. These modifications include:
- The observed column from the MTZ data.
- The SigF column from the MTZ data.
- The RFree validation column information from the MTZ data.
- The number of refinement cycles to run.
- The XRay Weight to use.
- The type of refinement to perform, ‘Normal’ or ‘Rigid’.
REFMAC5 Script¶
The REFMAC script can be seen in the Script tab of the widget. This tab is editable (except for the CYCLES, WEIGHT and MTZ columns).
Changes made to the script are remembered for the next run.
PHENIX Script¶
The PHENIX tab of the widget allows the user to choose between using the forcefield helper or a dictionary file.
Running REFMAC5¶
To run REFMAC5, simply click on the Refine button, or, if in manual mode, click on Save and run the script from the command line. The output of the refinement program appears in the Run tab of the window.
When successfully completed, the Next button will appear.
Running PHENIX¶
To run PHENIX, simply click on the Refine button, or, if in manual mode, click on Save and refine from the command line. The output of PHENIX appears in the Run tab of the window. PHENIX can be run in two modes; using AFITT helpers (MMFF only) or using the AFITT generated dictionary file.
When successfully completed, the Next button will appear.
Notes about the Refinement Dictionary¶
Dictionary generation uses the MMFF94 or MMFF94s forcefield to form geometric constraints for ligands and proteins under refinement. AFITT generates fully compliant residue dictionaries that encode these constraints.
When writing out a residue, AFITT checks to see if the residue name already exists in the published list of monomers [public-monomers]. This list has been validated against the list of monomers published with REFMAC5 and PHENIX since they are not completely identical.
If the residue exists and is a compatible match, the atom names of the input ligand are remapped to match the atom names of the public monomer.
If the residue is not a compatible match, a new residue is generated starting with residue name UNL and then progressing to OE1, OE2... and so on.
In all cases, a new residue dictionary is generated using the specified MMFF94 or MMFF94s forcefield to be used in refinement.