Preparing Apo Proteins as Model-Ready Protein Structures (October 20, 2023)
What you will learn: How to generate and prepare a design unit for an unliganded (apo) structure.
Identifying binding pockets on apo proteins can significantly enhance the scope of drug discovery. Spruce streamlines the process of detecting potential binding sites on unliganded protein structures.
The Spruce - Protein Preparation floe is available in Orion. Spruce is also available as a downloadable command-line application and as a programmer’s toolkit, offering three options for preparing design units (DUs) for apo proteins. The resulting DU includes the OEReceptor, which contains essential binding site information. You can choose one of these options:
Provide a reference DU or PDB code or file that identifies the binding site of interest.
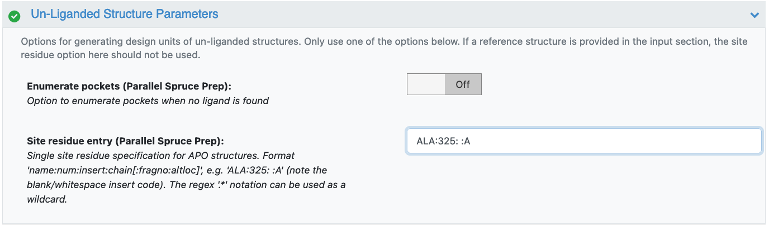
Provide a single residue in the binding site of interest (for example, HIS:102: :A).
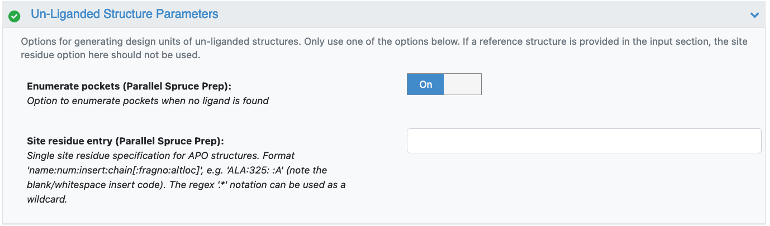
Enumerate potential binding sites. In this case, the floe sends prepared biological units to a pocket finding cube that will enumerate potential binding pockets into individually prepared design units.
Option 1: Provide a reference protein to identify the binding site on the apoprotein. Input a holo protein structure PDB (liganded) or its design unit as reference to find the binding site on the apoprotein.

Option 2: Add a site residue to identify the site of interest. Use the option below to insert a single residue to specify the region of interest.
Option 3: Enumerate potential binding sites. Toggle On if you would like to find pockets with
OEPocket and F-Pocket.