Filter Docked Poses Using Interaction Type (August 17, 2023)
When analyzing the results of a structure-based virtual screen, such as the results of Gigadock™ or Gigadock™ Warp in Orion, often the next step involves filtering out docked poses that do not exhibit specific interactions between the ligands and the protein. To simplify and automate this critical process, we have developed a floe named Filter Docking Hits by Interactions in Orion.
This floe requires two input datasets: 1) the docked poses of the ligands, and 2) the receptor that the ligands were docked into. The output is the receptor annotated with interaction type, and a dataset of the docked poses annotated with interaction hints. (Note: These interaction hints are hidden from the spreadsheet view by default and can be made visible via the Data Handling menu in Orion.)
To View the Docked Poses and Filter on the Interactions
Step 1: Make both output datasets from the Filter Docking Hits by Interactions floe active and navigate to the 3D page. Then add a filter (or multiple filters).
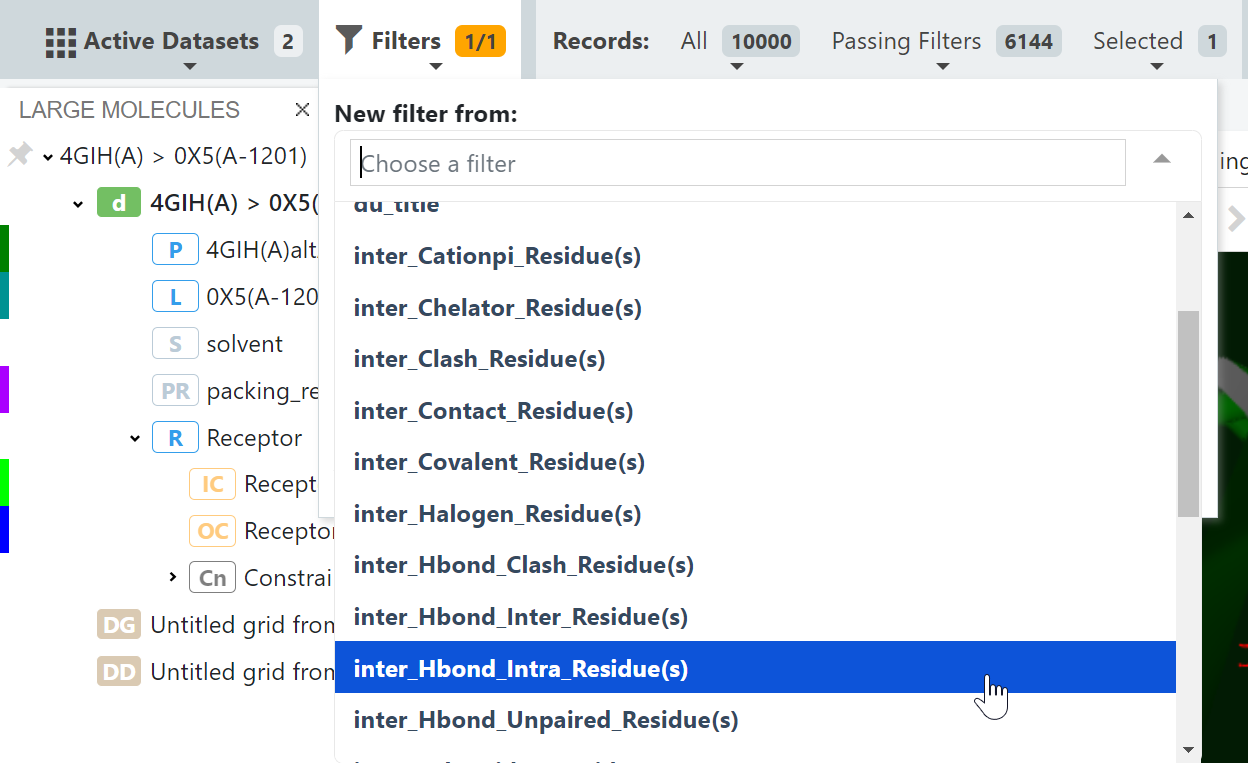
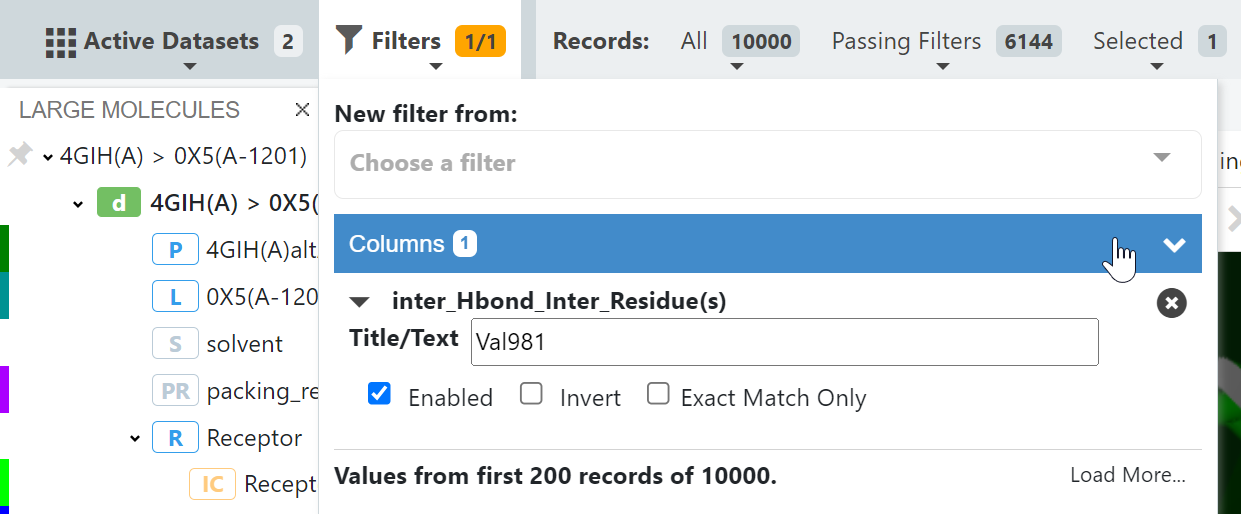
Step 2: Enter the desired intermolecular hydrogen bond to residue (Val981) pass the filter.
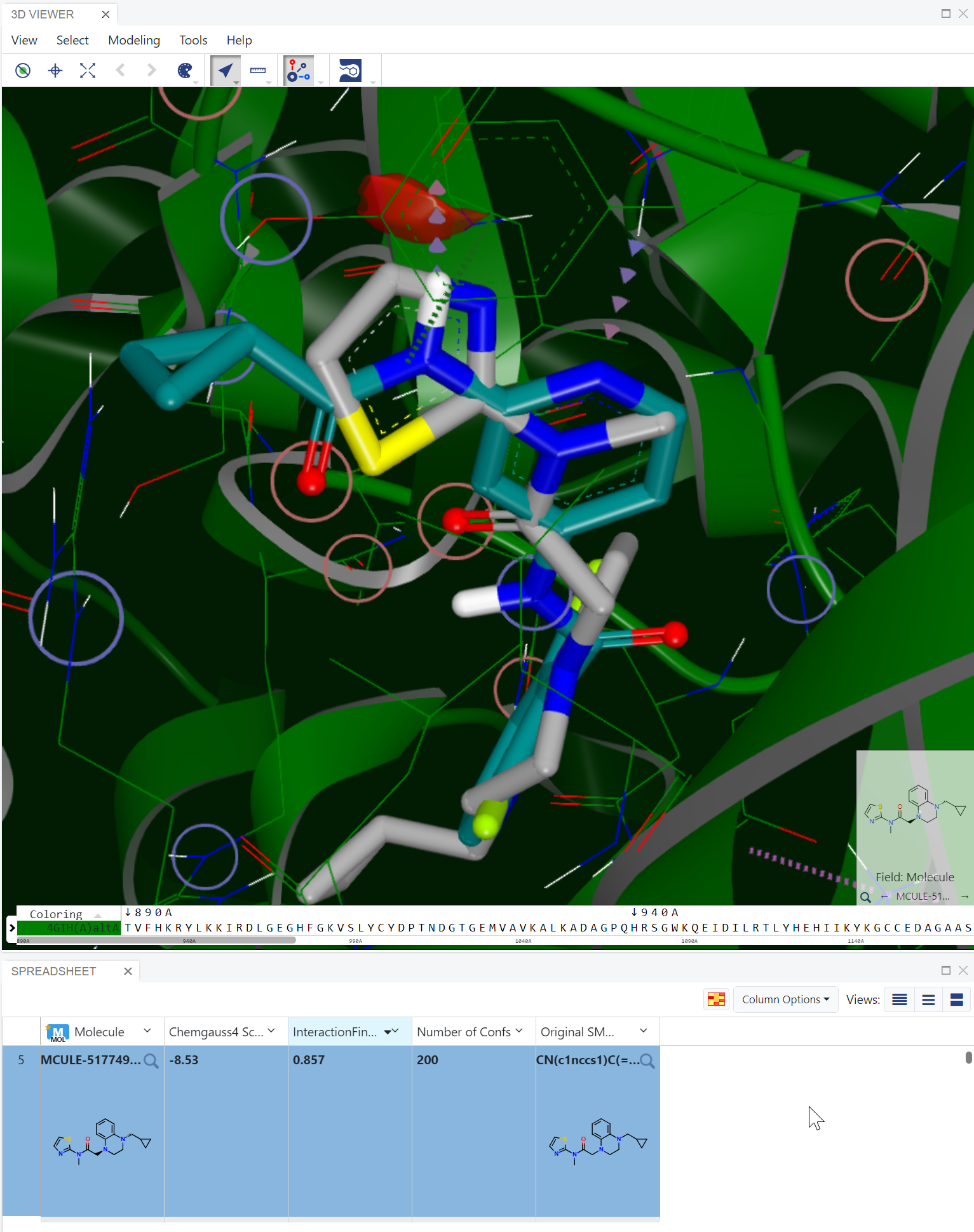
If the receptor contains a bound ligand, the floe provides an additional valuable feature by creating a field called “InteractionFingerprintSimilarity” in the output dataset, alongside the annotated docked poses. This similarity score is represented as a Tanimoto score ranging from 0 to 1, indicating the degree of similarity between the interactions observed in the docked ligands and those observed in the co-crystallized ligand. A high similarity score indicates that the docked compound has a binding mode like the co-crystallized ligand.
Step 3 (optional): Sort the docked ligand (carbon atoms shown in grey) by InteractionFingerprintSimilarity. A score of 0.857 indicates that its binding mode is very similar to the co-crystallized ligand (carbon atoms shown in dark green).