How to Extract Proteins, Ligands, and Other Components from Design Units (December 22, 2023)
What you will learn: How to extract proteins, ligands, and other components from design units.
Design Units (DUs) constitute an integral part of Orion, encompassing information about a protein, bound ligands, and other components like excipients and solvent. DUs serve multiple purposes in Orion, such as in docking and molecular dynamics (MD) simulations.
Occasionally, it becomes necessary to convert a DU into a molecule or a PDB file for specific analyses or sharing with colleagues. These tasks can be accomplished in Orion by executing either of these Floes:
DU to Mol Floe for conversion into a dataset.
DU to PDB Floe for conversion into a PDB file.
These Floes are versatile, allowing the conversion of single or multiple DUs. For instance, DU to Mol can be utilized to extract all ligands from the ligand-protein binding site search hit list, enabling the examination of ligand overlays without the contextual protein information.

Under the parameter setting Components to be part of the molecule, unselect all and set them to undefined.
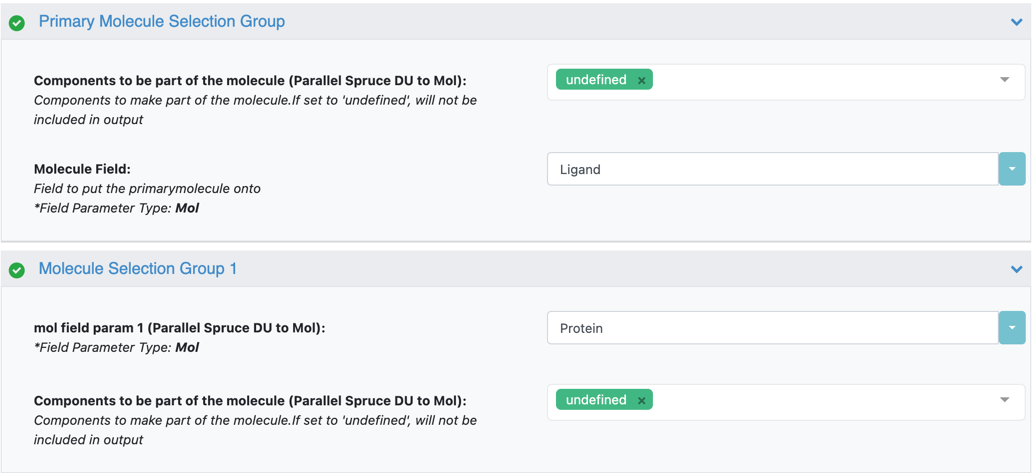
Setting the parameter to undefined ensures that only the ligand(s) will be included in the output.
Click on the link for more information on the DU to Mol Floe.
Likewise, the DU to PDB Floe converts a design unit into a PDB file. (Please note that the resulting file will be saved in My Data/Exports.) The only input required is a dataset containing one or multiple DUs.

For this Floe, you can customize the inclusion of DU components in the output by adjusting the Components to be part
of the molecule parameter in the Output Selection section. Click on the link to get more information on the
DU to PDB Floe.

Access more Orion Tips and Orion video tutorials.