OEToolkits 2015.Feb
This release of the OpenEye Toolkits contains many new features and bug fixes. The most important are highlighted here.
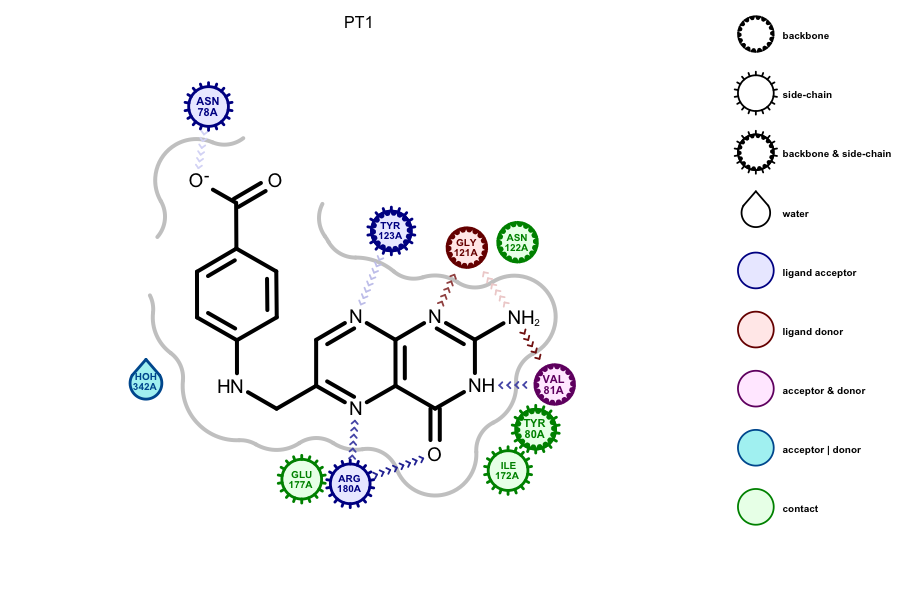
Protein-ligand Rendering
Grapheme TK now includes a powerful new API for the display of 3D information. The interactions between a protein and a ligand provide crucial information to the pursuit of new drugs. To facilitate the deciphering of these complex interactions, OpenEye is releasing the ability to render this 3D information in a 2D diagram. Currently, only OEDocking TK interactions can be rendered by Grapheme TK. However, this feature will be fully extensible through the OEBio TK. If you wish to display a particular type of interaction and would like to contribute to the development, please contact support@eyesopen.com.
Documentation Refresh
To improve usability and maintain a high level of quality, the OpenEye Toolkit documentation has been refreshed over the last year. In addition to a new logo and style, significant functionality has been added. All OpenEye Toolkits will now cross-reference, leading users to the correct toolkit while searching for functionality. In addition, searching the HTML documentation can now be performed with Google, offering significantly better search results than the previous HTML documentation. If you are an avid user of the PDF documentation, we strongly encourage you to take another look at the HTML documentation.

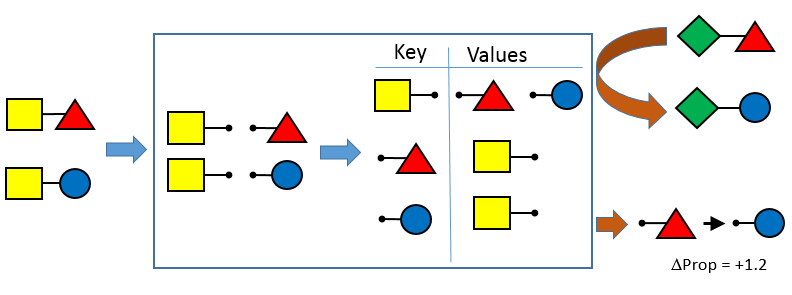
Matched Pairs Transformations
This release continues the incorporation of community suggestions in the OEMedChem TK. In this release, the matched molecular pairs functionality has been augmented to include easy access to the transformations themselves in native form, allowing for retrieval and manipulation of transformations for many more matched pair use cases.
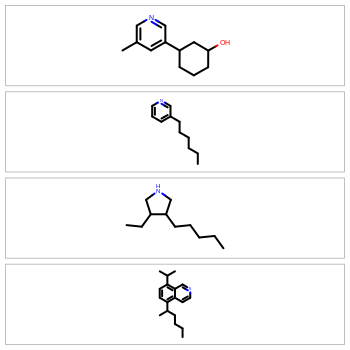
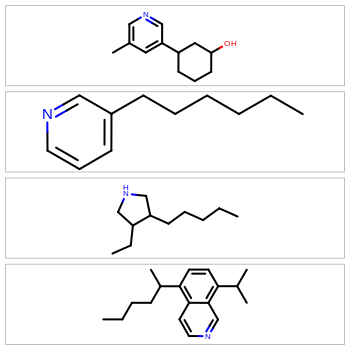
Optimized Depiction Orientation
Molecular depictions are a common element in any chemistry environment. However, they can waste a lot of screen real estate, especially when displaying extended or oddly shaped molecules. To maximize screen space, the OEDepict TK now provides the ability to optimize for either horizontal or vertical rendering of the molecule coordinates. For example: to display molecules in a spreadsheet, horizontal depictions will conserve the most vertical whitespace; to display molecules in the sidebar of a webpage, vertical rendering will conserve the most horizontal whitespace.
|
|
General Notices
SuSe 11 is no longer supported.
Visual Studio 2008 for C++ and C# toolkits is no longer supported.
The 2015.Jun release will be the last to support Visual Studio 2010 for C++.
OSX 10.10 Yosemite support has been added.
Babel is a command line tool for molecular file format translation, incorporating a wide range of OEChem options. The application code is now open source and can be found here: https://github.com/oess/OEBabel
OpenEye Python toolkits are now built with SWIG 3.0.2. Users who write and compile their own OpenEye C++ toolkit extensions will need to upgrade accordingly. An example of compiling a custom extension to the Python toolkits is given here: https://github.com/oess/oepython_extension.