OEToolkits 2019.Apr
FastROCS TK: Color calculation on GPU
Changes to the FastROCS algorithm, including porting the static color calculation to GPU, have resulted in a more than 2x performance improvement of FastROCS on some GPU systems. Benchmarking was carried out on AWS GPU instances and compared to the 2018.Oct version of FastROCS TK:
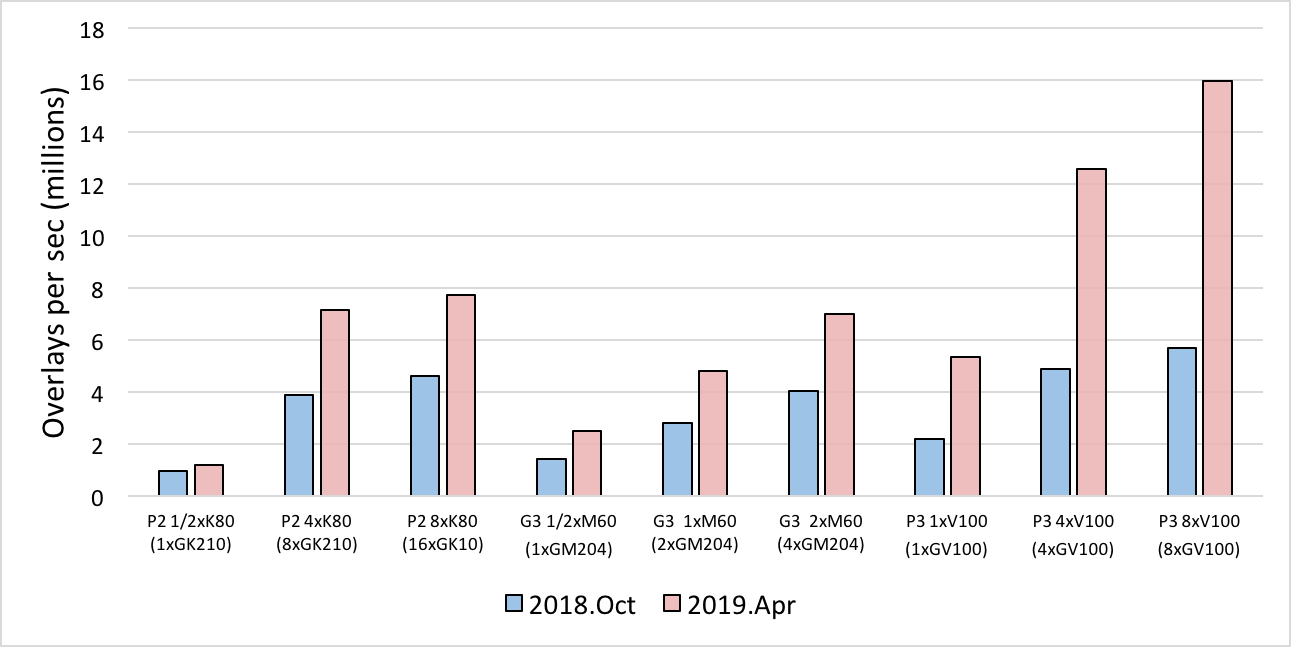
Performance comparison between the 2018.Oct release and the 2019.Apr release using OpenEye’s standard benchmark dataset of 14M conformers on AWS.
For some systems, the standard benchmark dataset of 14M conformers is not large enough to fully saturate the GPUs. To demonstrate the potential performance of larger databases, a 2 billion conformer subset of Enamine was searched on suitable AWS instances. FastROCS searched the 2 billion conformer database on an AWS P3 instance with 8x Nvidia Tesla V100 GPUs at a rate of over 44 million overlays per second:
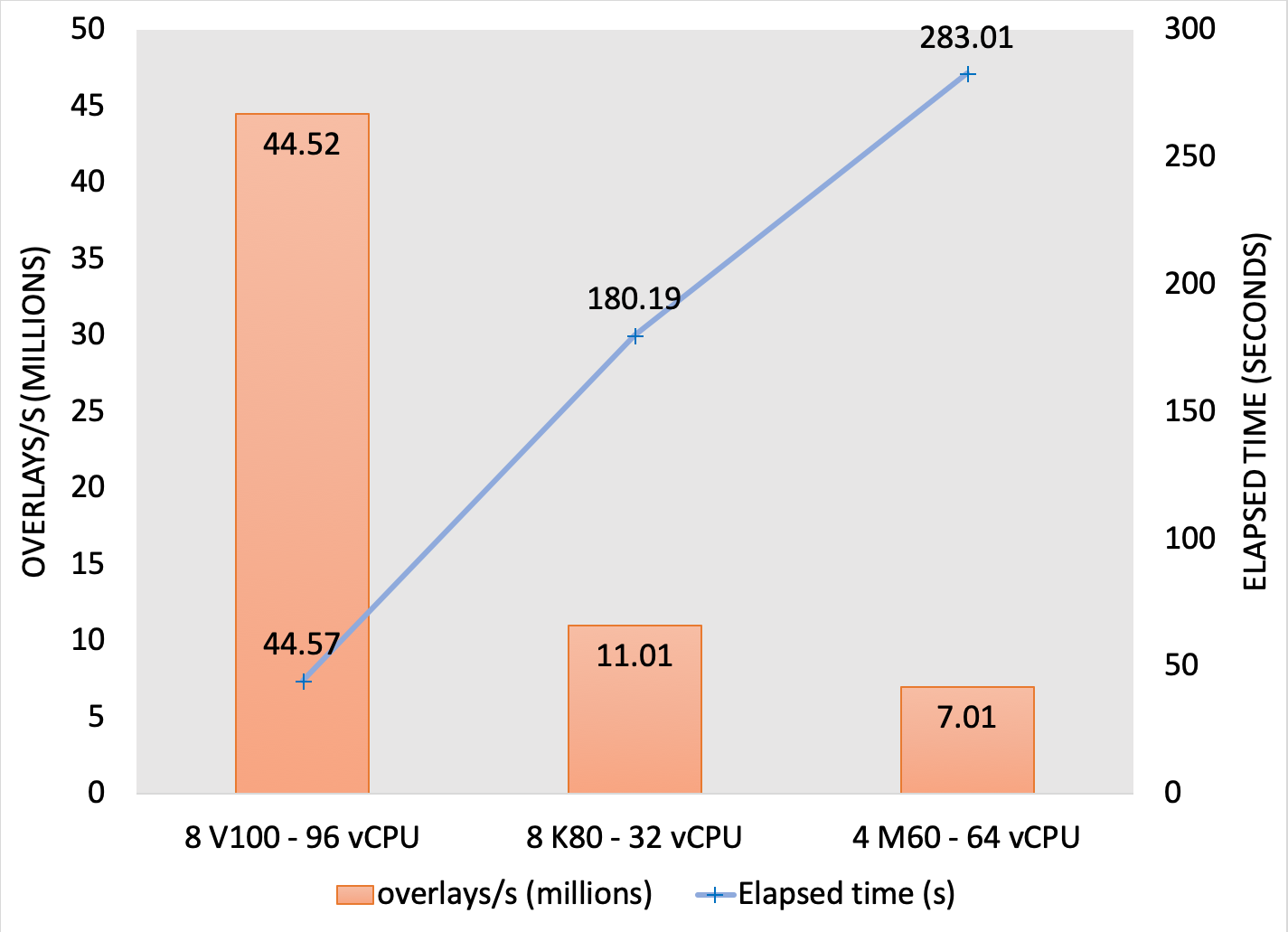
Performance of a 2 billion conformer subset of Enamine on suitable AWS instances.
OEChem TK: OEZ compressed file format
A new compressed file format with the extension oez has been added to
OEChem TK.
This format uses the Zstandard algorithm
to efficiently compress individual multi-conformer molecules, making it well-suited
to work with OEMolDatabase for fast read-only random access to molecules.
See more details in the Compressed Input and Output section of the
OEChem TK documentation.
OEGrapheme TK: Ramachandran plot
Following our tradition of providing easily comprehensible visualization of complex data, OEGrapheme TK can now depict Ramachandran plots ([Ramachandran-1963]). These plots provide an easy way to examine the distribution of backbone dihedral angles in a particular protein. For instance, structure quality can be assessed by identifying residues whose backbone \(\psi\) are outside energetically accessible regions. Grapheme TK uses the data and classifications available in OEBio TK, which were extracted from the open-source Computational Crystallography Toolbox [Grosse-Kunstleve-2002].
Hovering over the red circles (on the top right plot) reveals the name of the outlier residues
Examples of visualizing the Ramachandran plots of various residue types in Grapheme TK (PDB: 2ROX)
General Notices
OpenEye currently uses OpenJDK8 to build its Java packages. We are currently exploring support for OpenJDK 11.
The 2019.Apr release skips Java support for macOS. For Java support on RHEL6, please contact support@eyesopen.com.
This release adds support for macOS 10.14. OS X 10.11 is no longer supported.
This release adds support for Ubuntu 18. Ubuntu 14 is no longer supported.
Visual Studio 2017 support has been added for C++ and C#. This is the last release to support Visual Studio 2013, as well as 32-bit Windows for C++ and C#.
Full support has been added for Python 3.7. Python 3.5 for Windows is no longer supported.
The following prerequisites have been introduced so that OpenEye can implement algorithms in a more modern and efficient C++11 style:
GCC 4.8 is now the oldest targeted compiler for toolkits. RHEL6 with the default GCC 4.4 compiler is no longer supported.
The minimum recommended GNU binutils version is 2.26 for the C++ single-build Linux packages.
The minimum CMake version required to build the C++ examples is now 3.1.3 on all platforms.