OEToolkits 2016.Oct
With this release, OpenEye continues to focus on protein handling and visualization. In addition, numerous improvements to the toolkits have been made.
Protein Preparation
A new function, OEPlaceHydrogens, has been introduced to the
OpenEye protein preparation workflow in OEBio TK. This function positions
hydrogens in a molecule to build hydrogen bonding networks and avoid clashes.
It can also flip functional groups that are sometimes modeled incorrectly,
such as histidine, asparagine, and
glutamine sidechains. Previously, OpenEye recommended that users run
the open source application Reduce to
perform these functions. OEPlaceHydrogens is easier
to use and has broader applicability because it uses SMARTS patterns rather
than relying on naming conventions.
This release represents an initial implementation of this functionality; it is under continuing development. A new example program, Preparing a Protein, demonstrates this functionality.
For more information, see the Protein Preparation section.
|
|
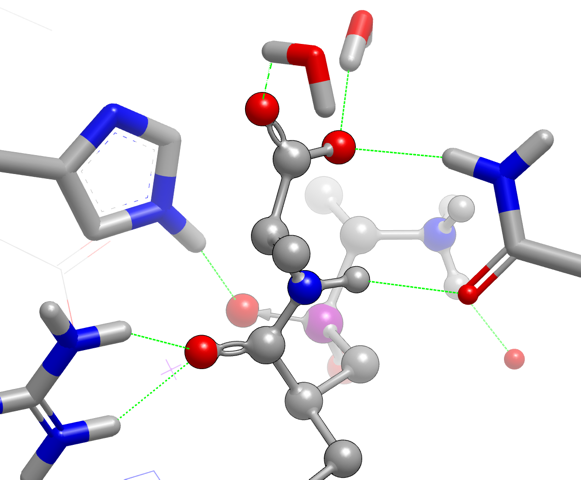
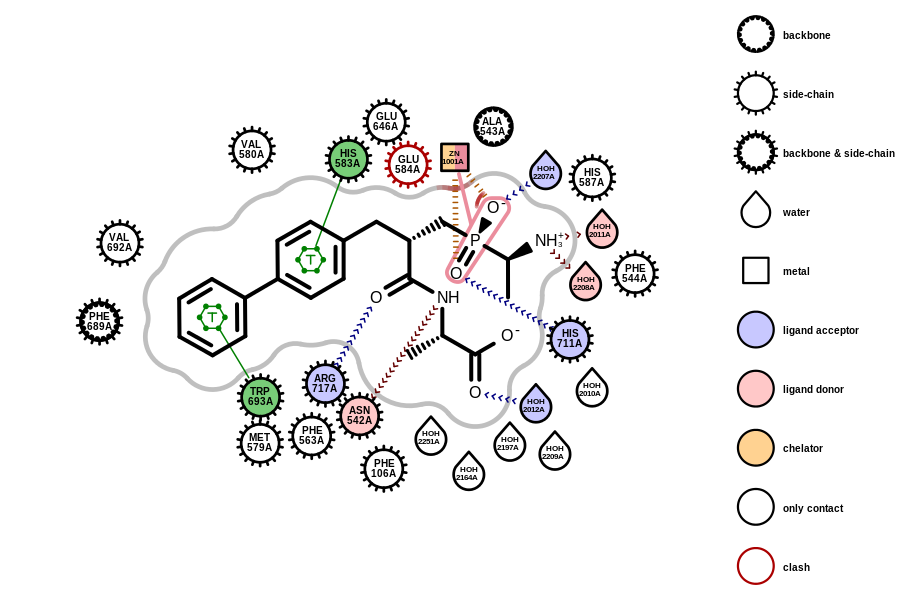
Protein-Ligand Interaction
This release of OEBio TK includes significant improvements to the perception of potential protein-ligand interactions, intramolecular interactions, and missing potential interactions.
A new type of interaction, Halogen Bonds,
has been added to OEBio TK.
It joins Hydrogen Bonds,
Salt Bridges,
Chelation,
and Pi- and T-aromatic Stacking
in the set of perceived interaction types. These interactions
provide qualitative hints about biological interactions that may be important
for binding, and can be helpful for visualizing interactions and clashes.
An interface for modifying the geometric definitions for each interaction has also been introduced. While the default bond geometries have been carefully constructed using literature data (see below), users can now customize interaction parameters to fit their needs.
Kumar, S. and Nussinov, R., Biophysical Journal, 83:1595-1612, 2002.
Cavallo, G., Metrangolo, P., Milani, R., Pilati, T., Priimagi, A., Resnati, G., Terraneo, G., Chem. Rev. 2478-2601, 2016.
Bissantz, C., Kuhn, B., Stahl, M., J. Med. Chem., 53:5061-5084, 2010.
Marcou, G., Rognan, D., J. Chem. Inf. Model., 47:195-207, 2007.
FastROCS TK CUDA Implementation
The internal GPU implementation of FastROCS has been ported from OpenCL to CUDA. Although this does not change the results returned by FastROCS and should not impact the deployment of FastROCS TK on customer machines, it does allow OpenEye to better support NVIDIA hardware and facilitates our ability to improve performance and add GPU functionality.
General Notices
The 2016.Oct release adds support for Ubuntu16 for C++, Java, Python 2.7, and Python 3.x.
The 2016.Oct release is the last release to support OSX 10.9. The 2017.Feb release will add full support for macOS Sierra 10.12.
Note
OpenEye continues to monitor and collect feedback from our customers on the need to support Python 2 wrappers. Our current plan is to phase out support for Python 2 on Windows by the 2017.Oct release and phase out support on other platforms at a later date. As this is a substantial change for our customers, we are willing to help with migration. Please contact support@eyesopen.com for more details.