Tutorials¶
These tutorials show how to use Floes and other OpenEye tools to generate and evaluate drug candidates starting from a PDB code.
Floes and Tools used in the Tutorials¶
The Classic Spruce Prep from PDB Codes Floe.
The Generative Structure Floe.
The Classic OMEGA Floe.
The Assign Partial Charges Floe.
The Classic Posit Floe.
The Short Trajectory MD with Analysis Floe.
The VIDA.
Prepare Protein and Bound Ligand from PDB Codes¶
An extracellular signal-regulated kinase 1/2 inhibitor is used in the tutorials. The PDB code is 5K4J and it is straightforward
to prepare the protein and bound ligand using the Classic Spruce - Prep from PDB Codes Floe.
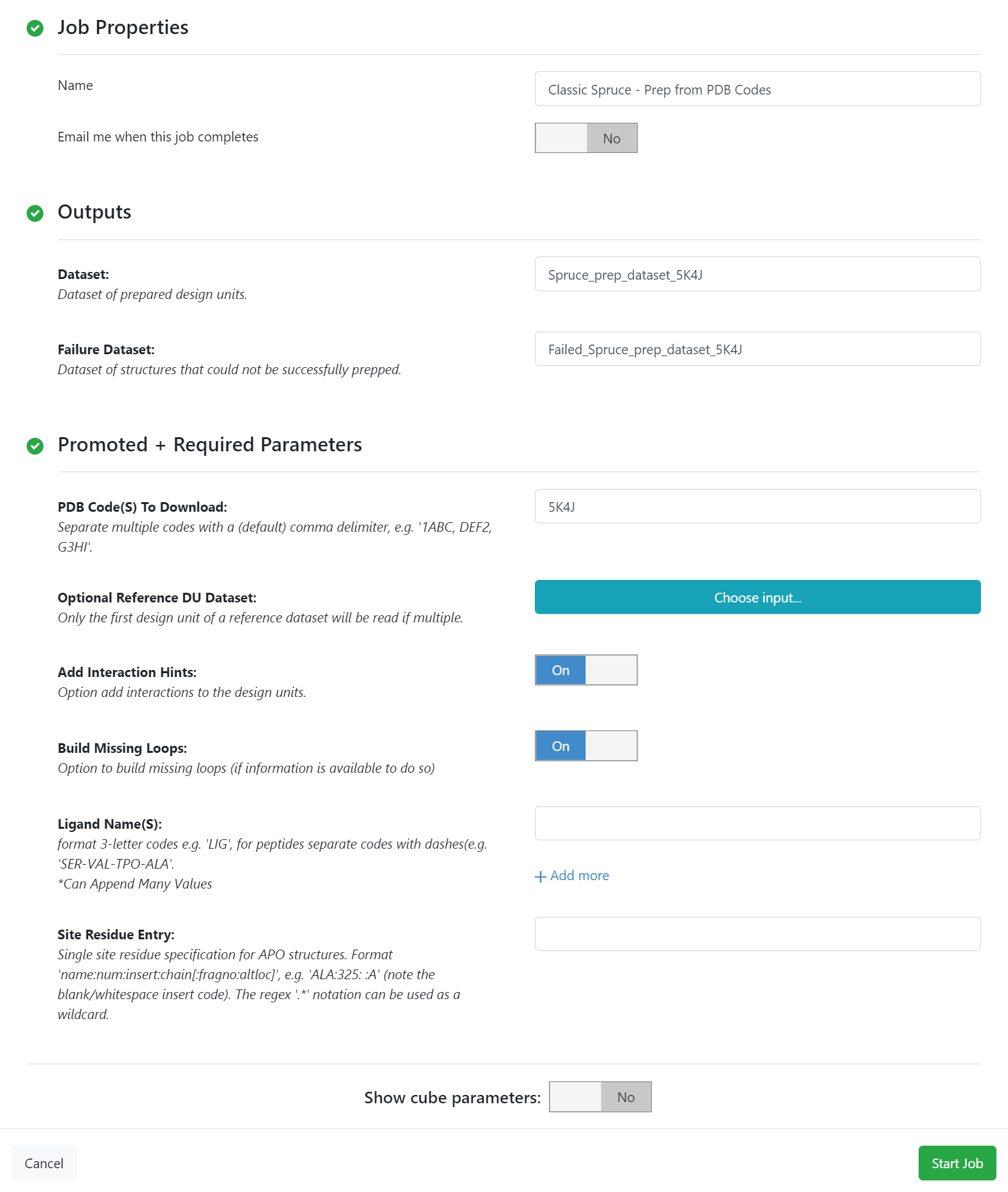
Spruce Floe Parameters¶
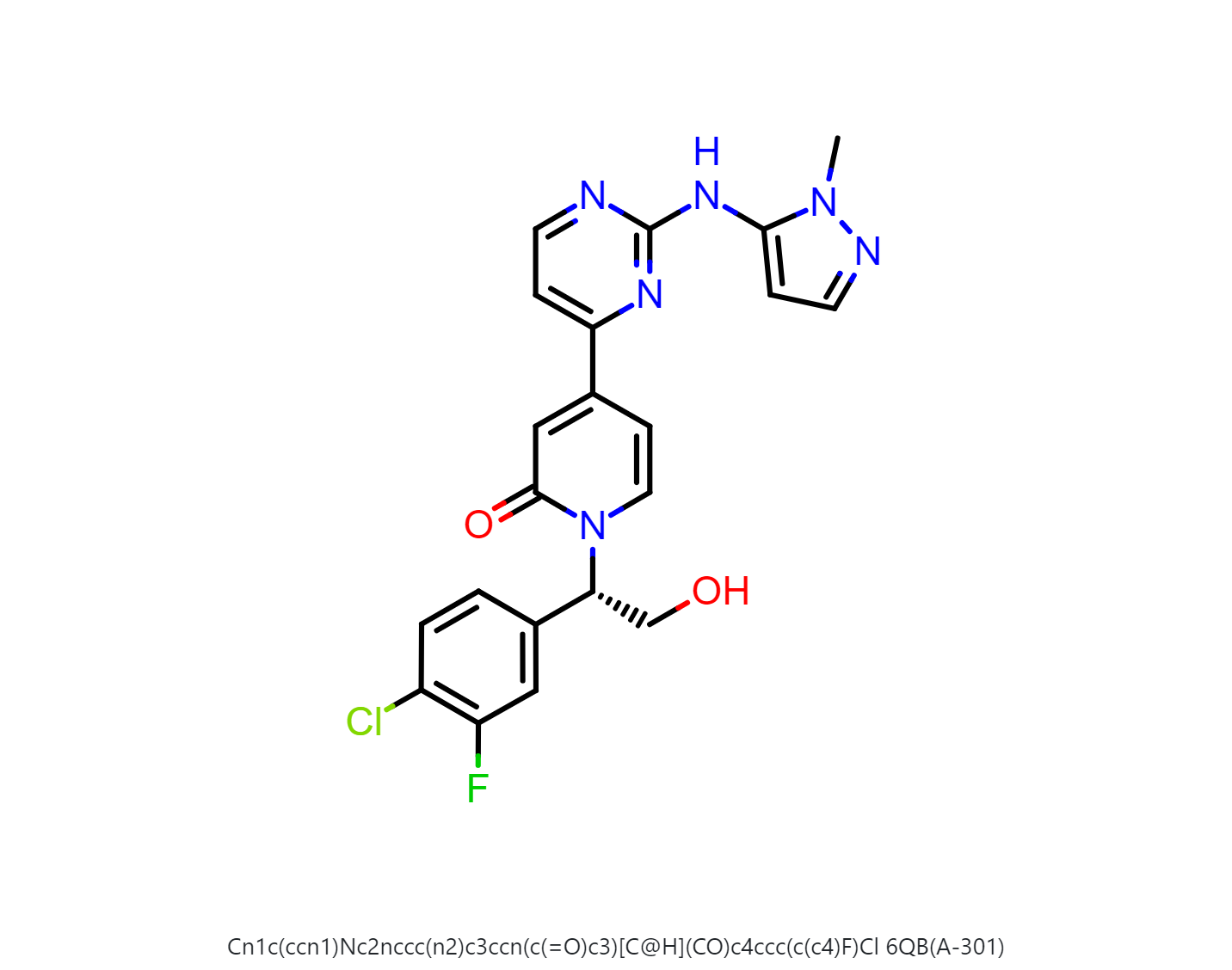
The output dataset Spruce_prep_dataset_5K4J needs to be downloaded as a oeb file and be loaded to VIDA to extract the bound ligand. The bound ligand (A-301) is uploaded to Orion as a dataset.
Generate and Prepare Candidates¶
The Generative Structure Floe is used to replace atoms in the bound ligand to create new drug candidates.
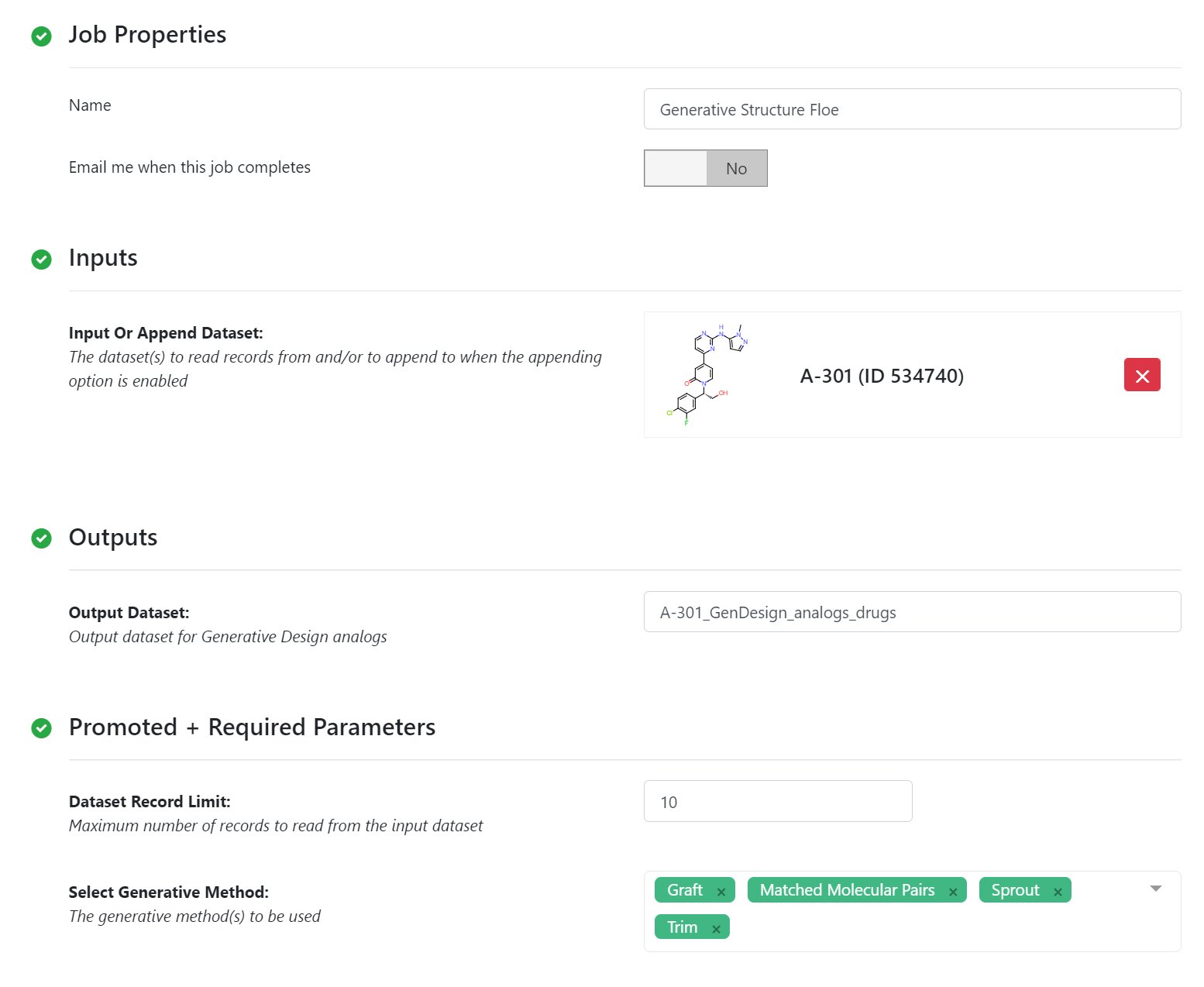
Generative Structure Floe Parameters¶
By default, the floe will generate 50-500 output structures for each input molecule. We can select Drug in Mol Filter dropdown list to only output drug like structures. The following figure shows the bound ligand and nine structures from the floe.
|
|
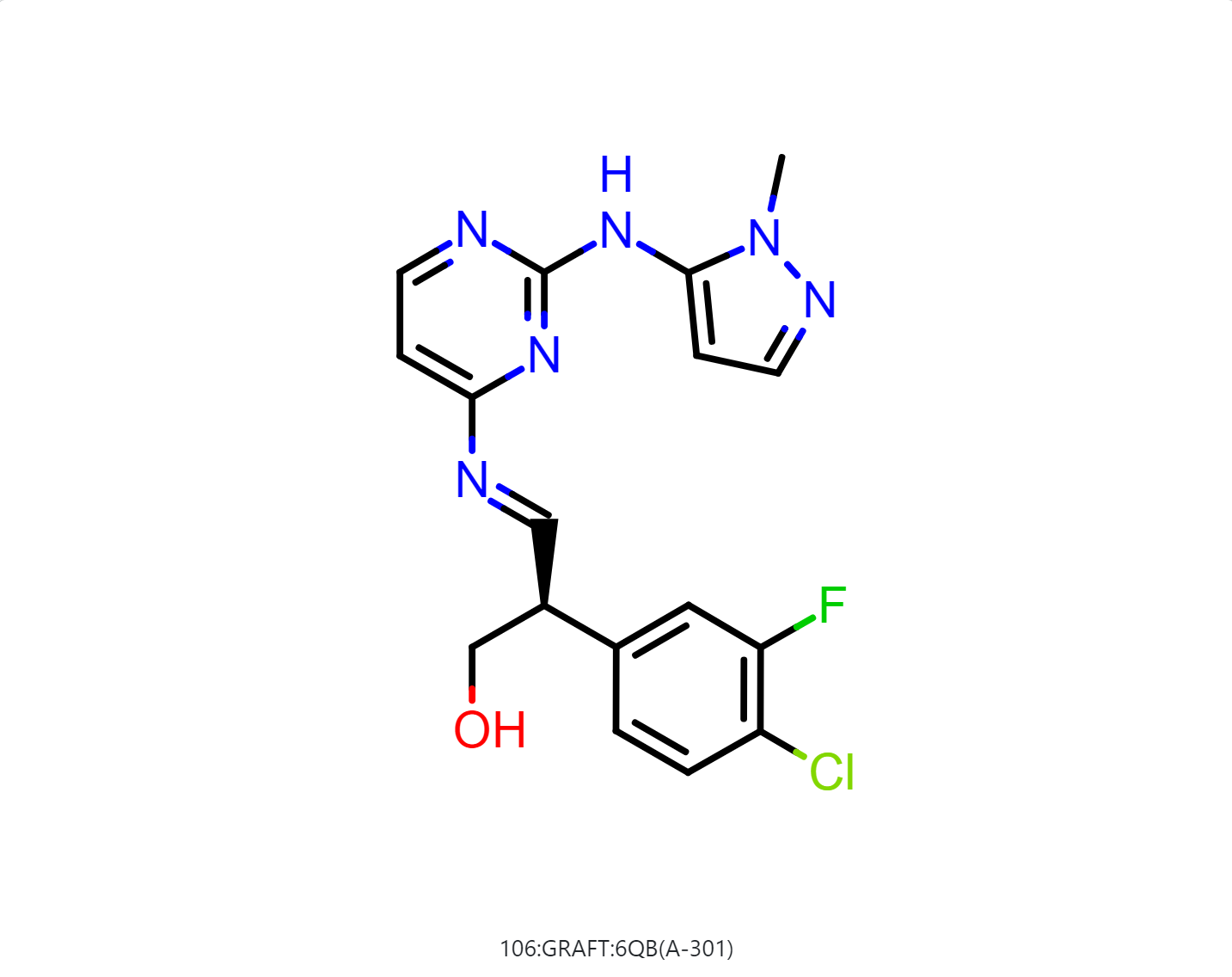
|
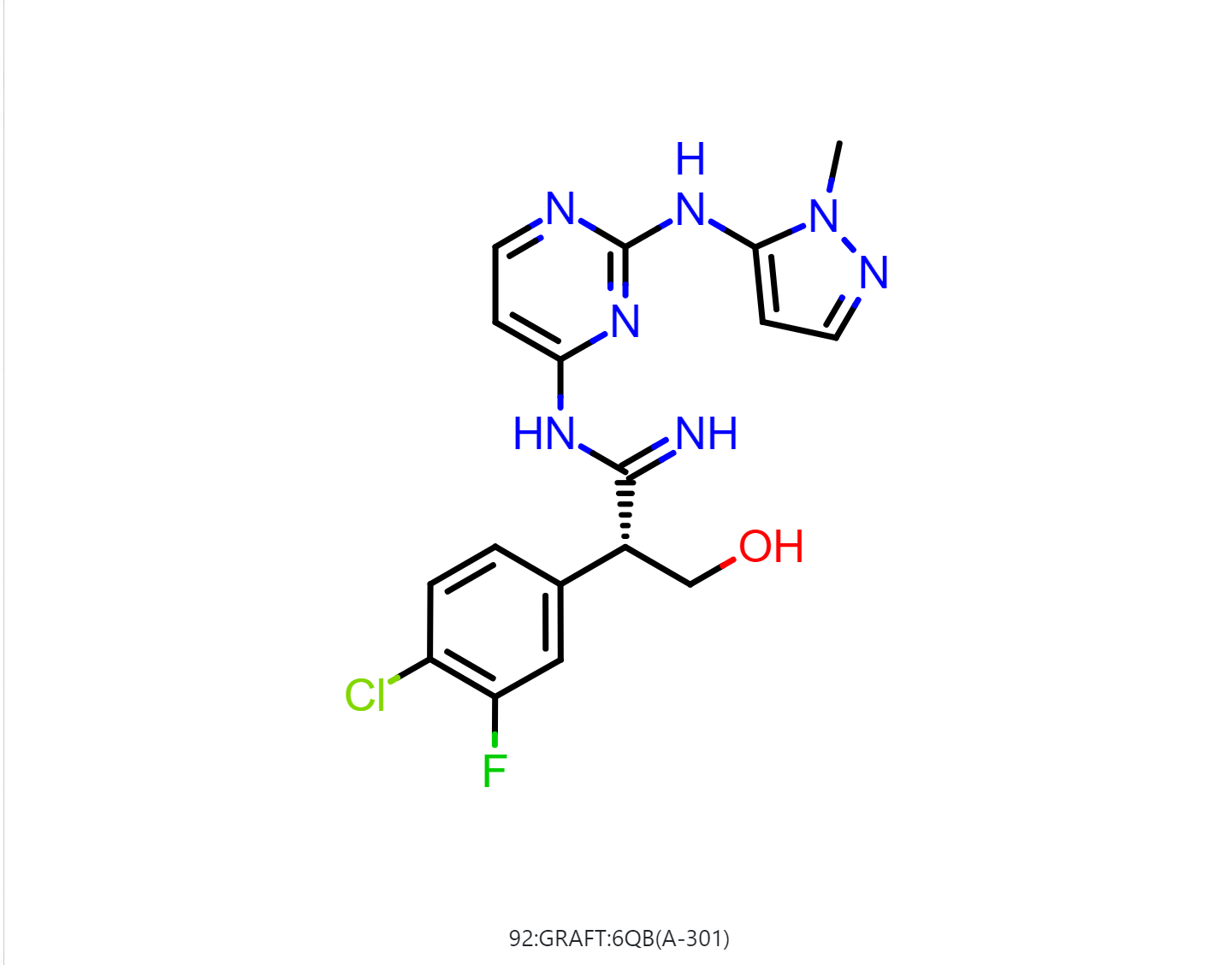
|
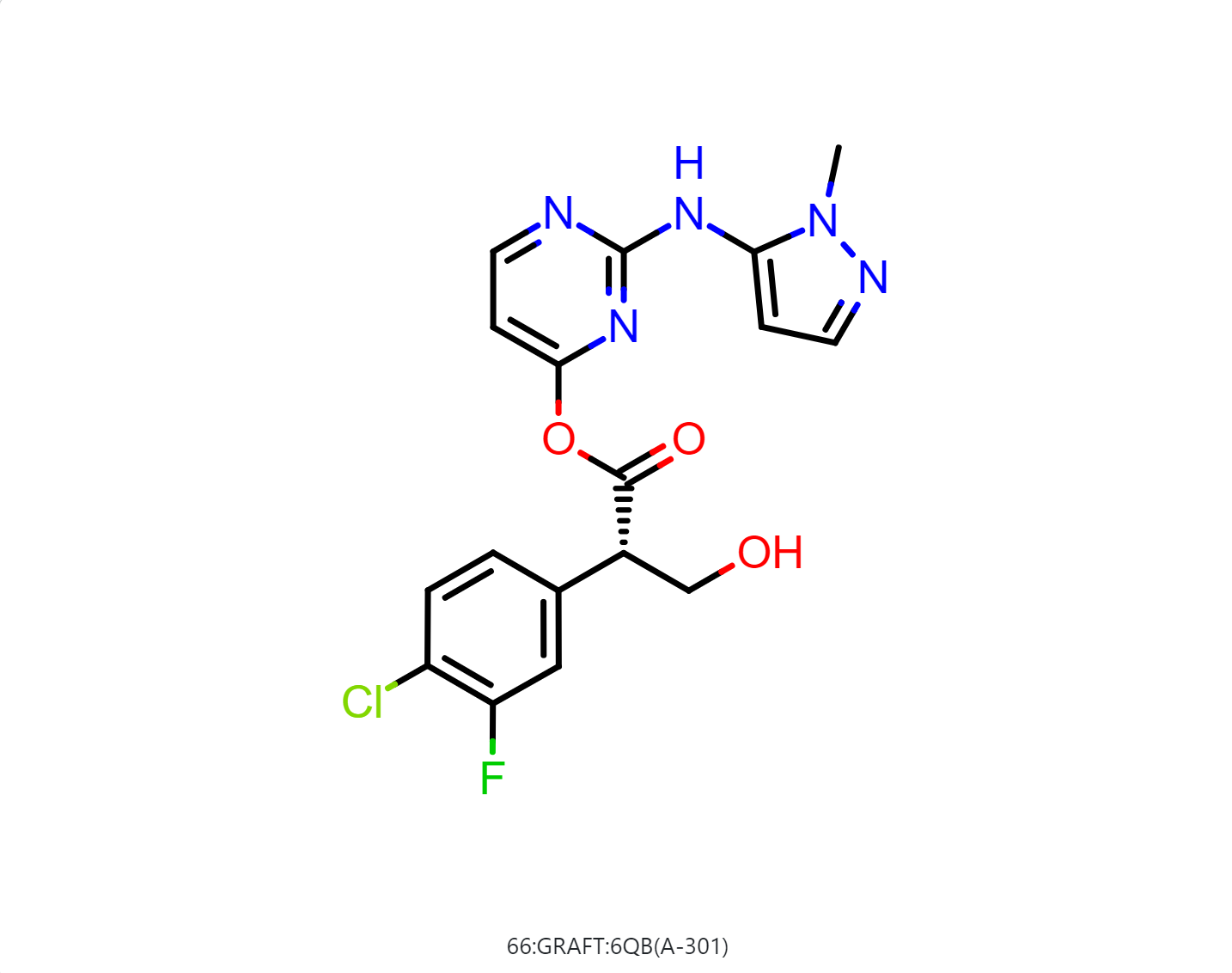
|
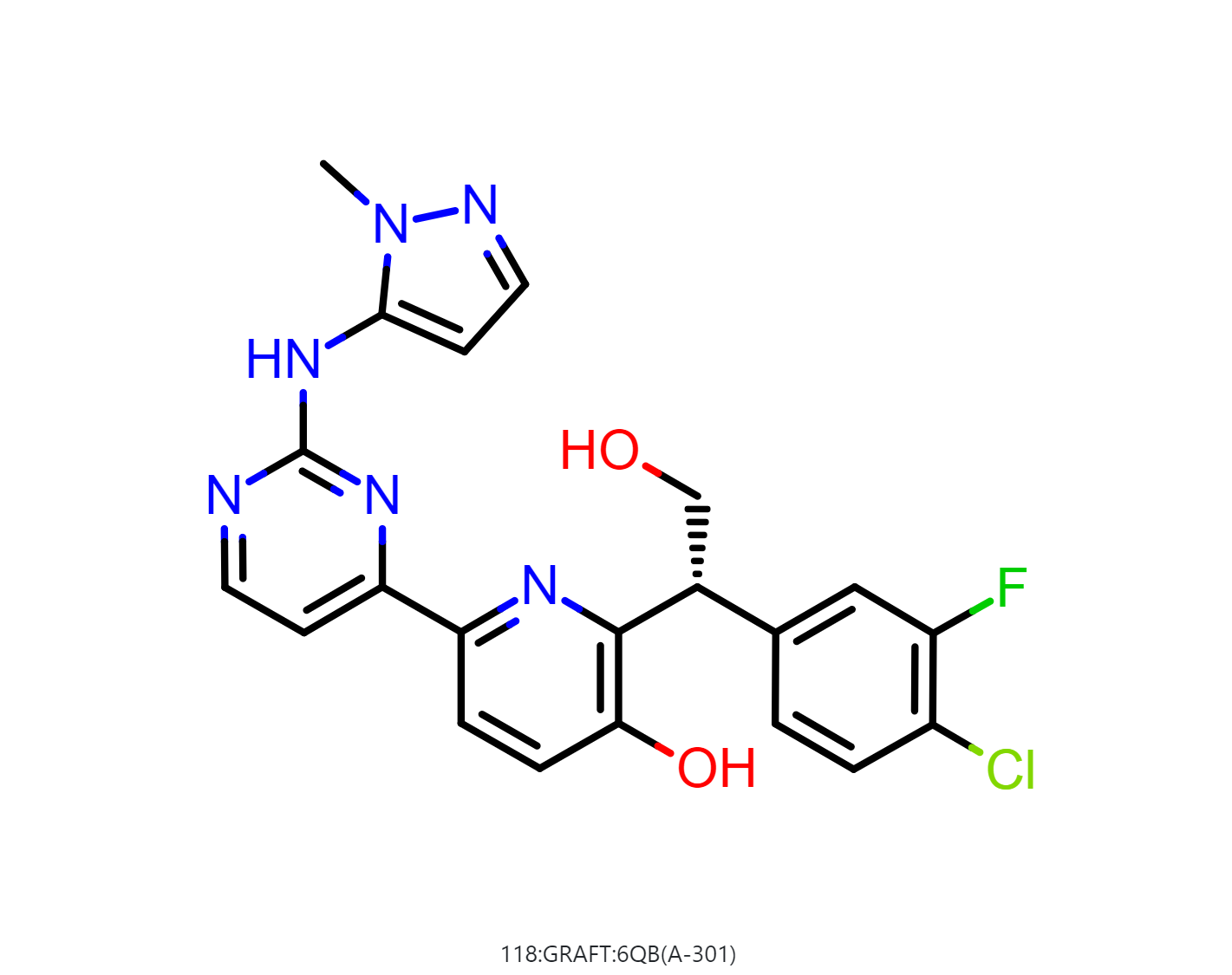
|
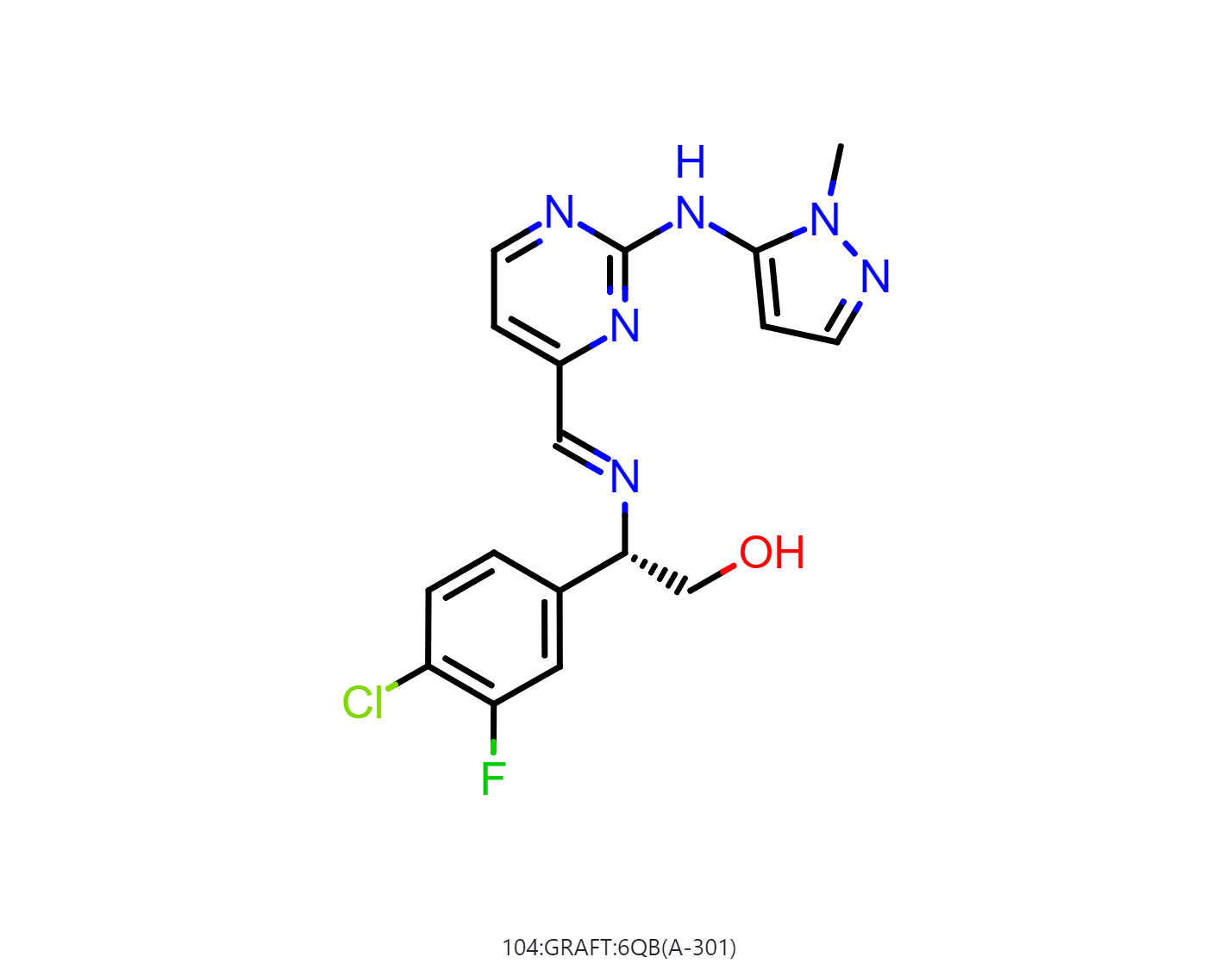
|
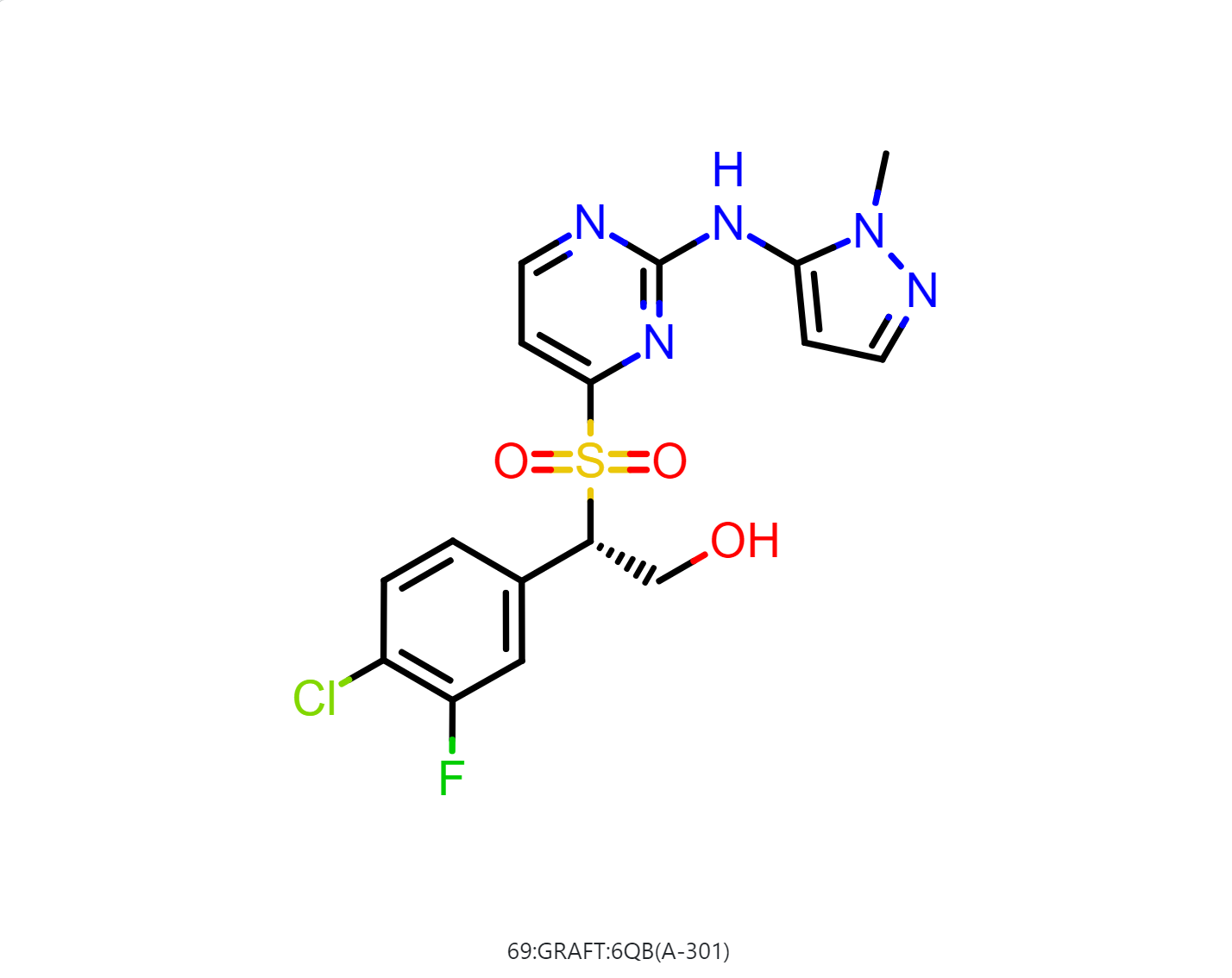
|
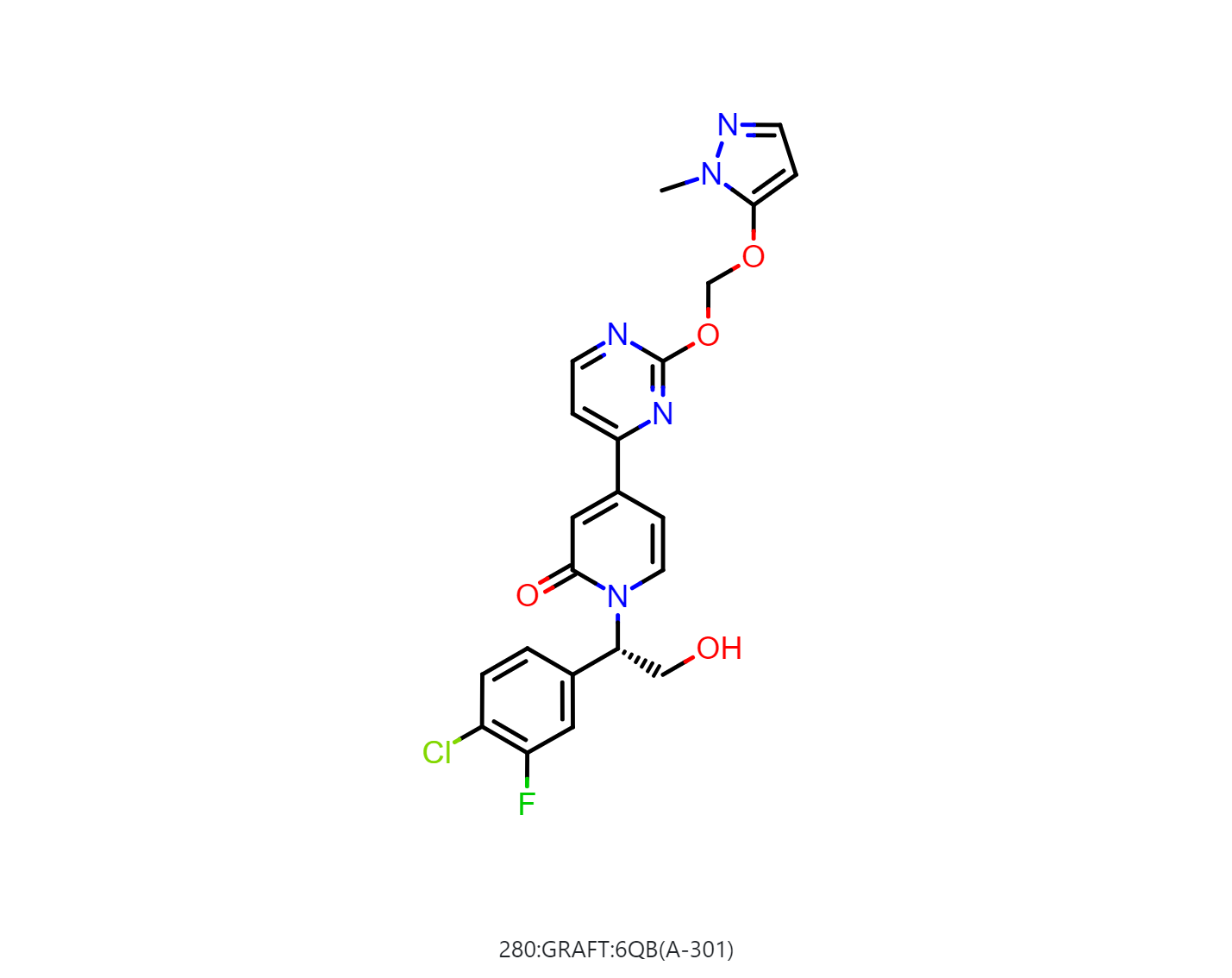
|
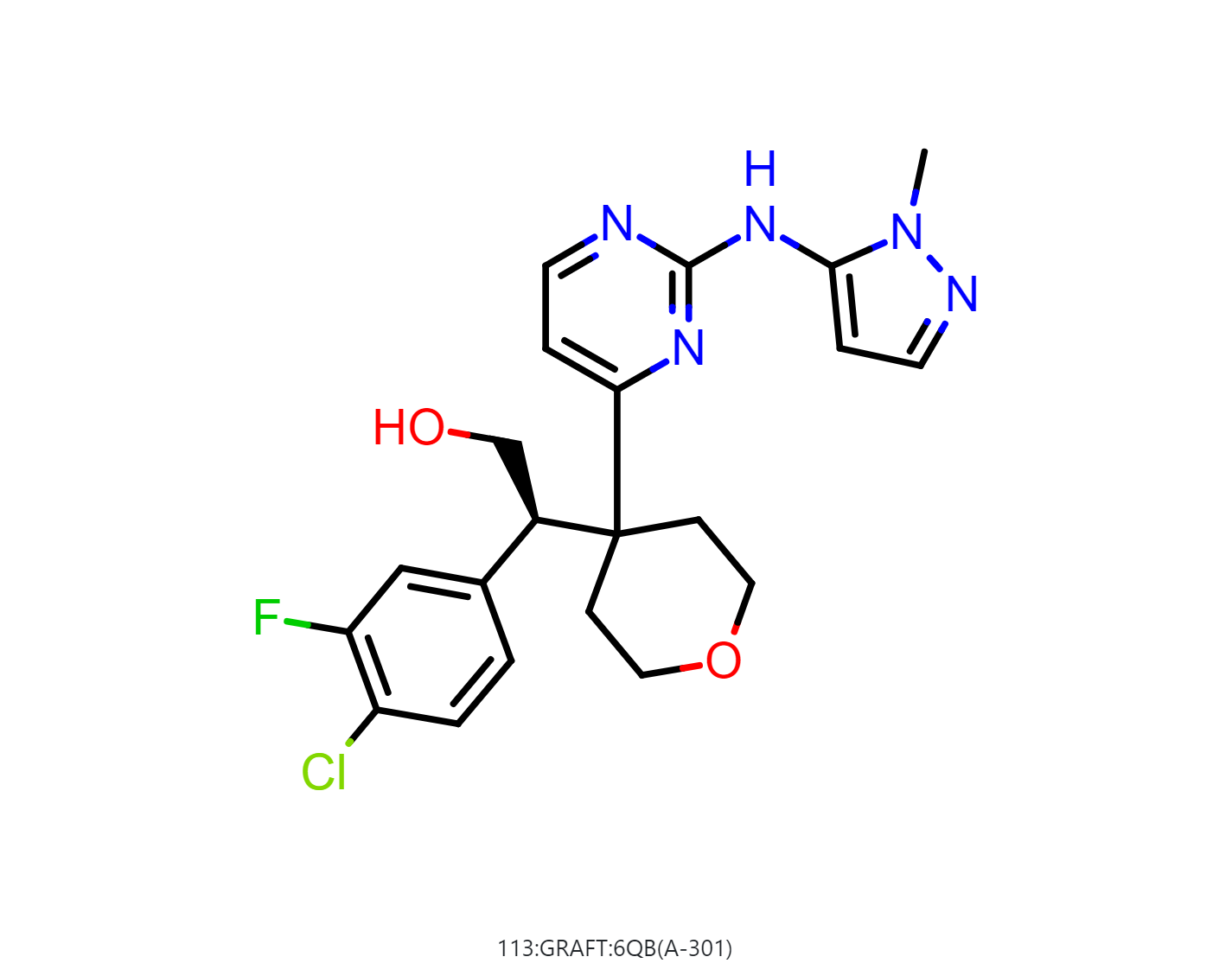
|
The 3D conformers of nine candidates are generated by the Classic Omega Floe using defualt parameters.
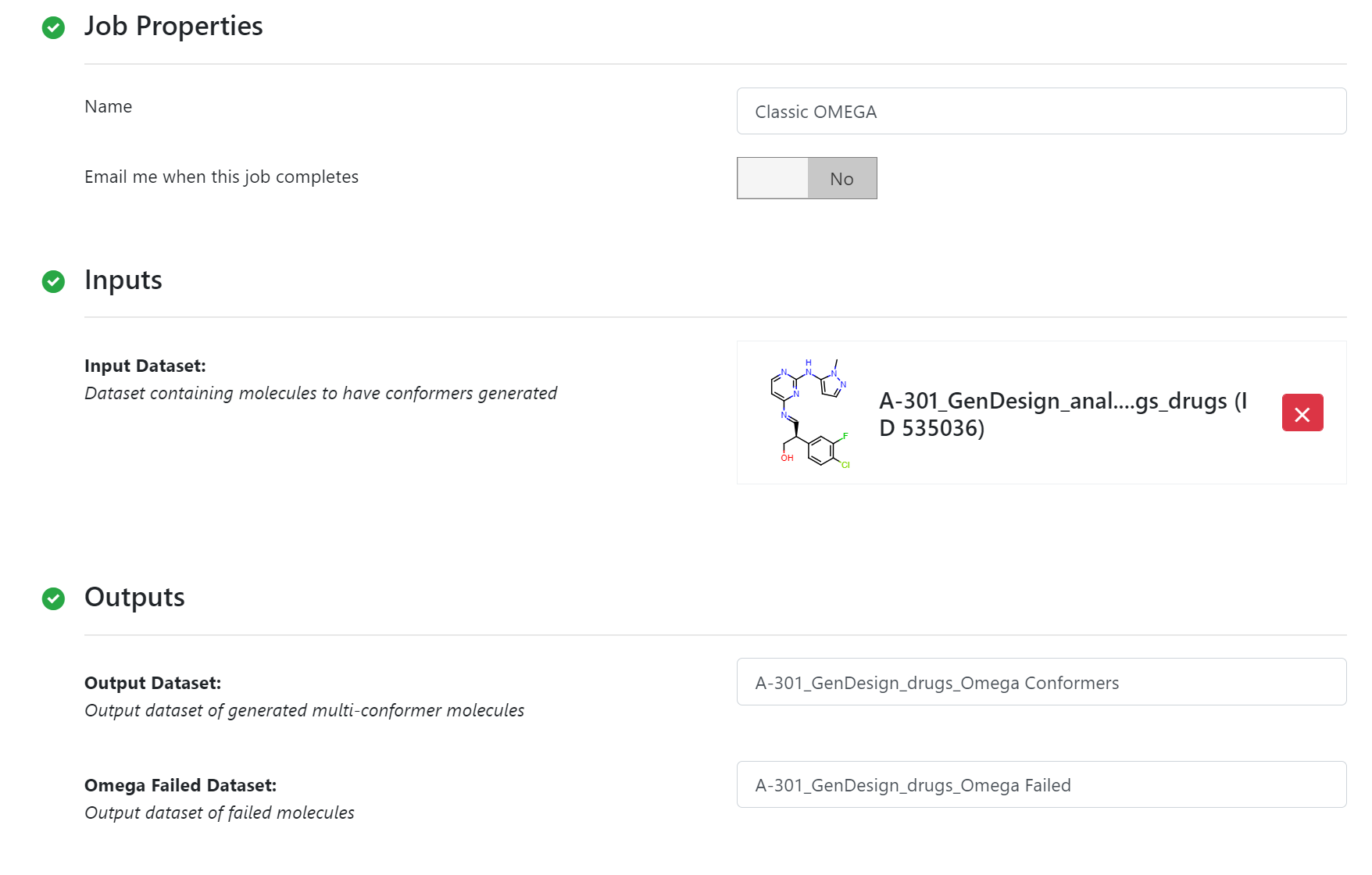
Classic Omega Floe Parameters¶
The final step of candidate preparation is charging. AM1BCCELF10 is used to charge the molecules, which allows up to 10 diverse conformers to be selected from those having the Electrostatically Least-interacting Functional groups (ELF). These conformers are then charged with the AM1BCCSym method and the charge sets are averaged to come up with a single charge set that is applied to all conformers. This yields good quality charge sets even for charged molecules.
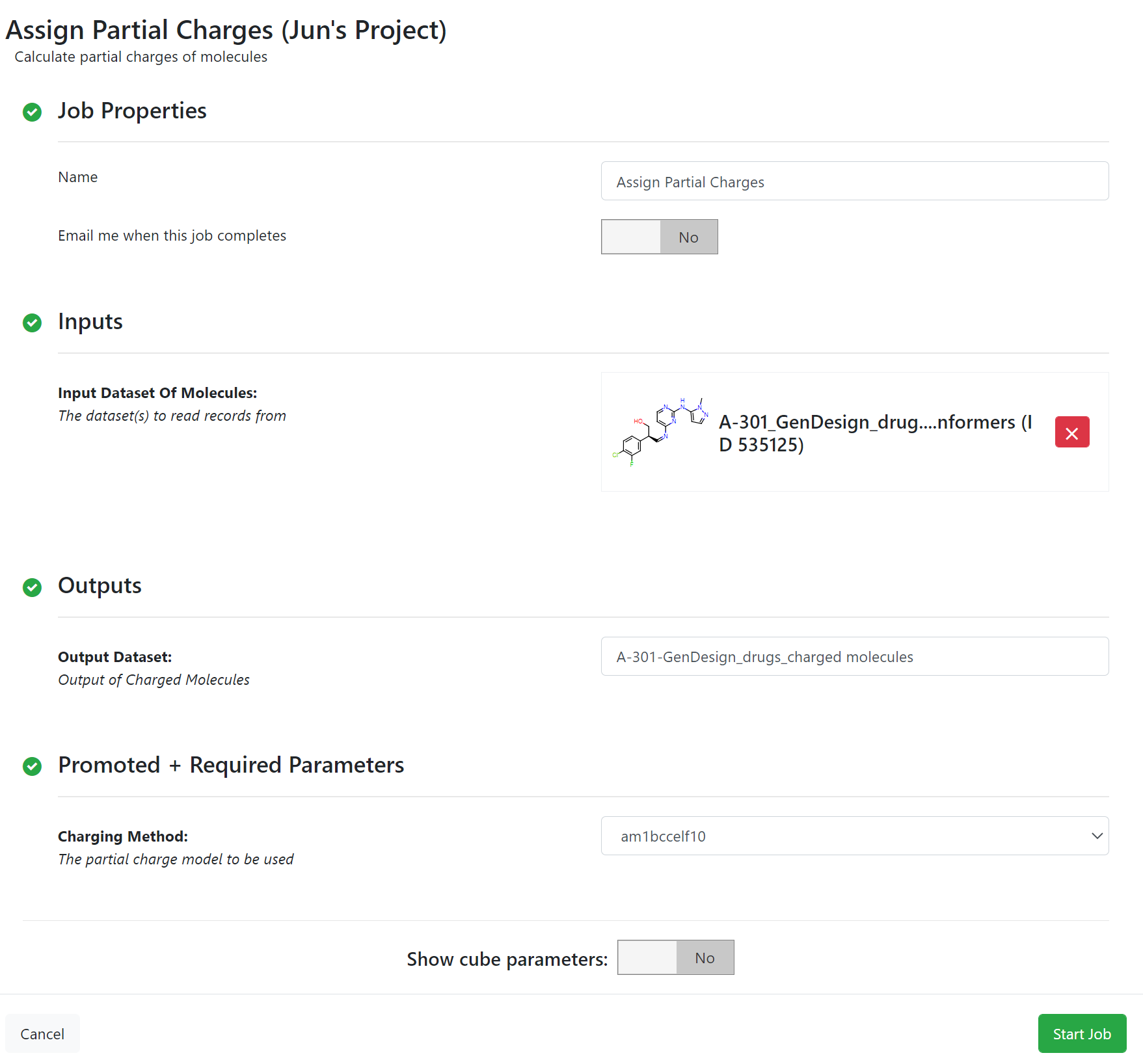
Partial Charge Floe Parameters¶
Screening of the Candidates using Docking and MD¶
The Classic Posit poses multiple ligands against multiple receptors, and it works best when fitting to a collection of co-crystal ligands, however, it can be used for a single ligand targets. Each generated pose is analyzed with a simple heuristic and marked with a probability. The charged candidaes and the Spruce_prep_dataset_5K4J dataset are used as inputs and default values of parameters are used to pose these candidates.
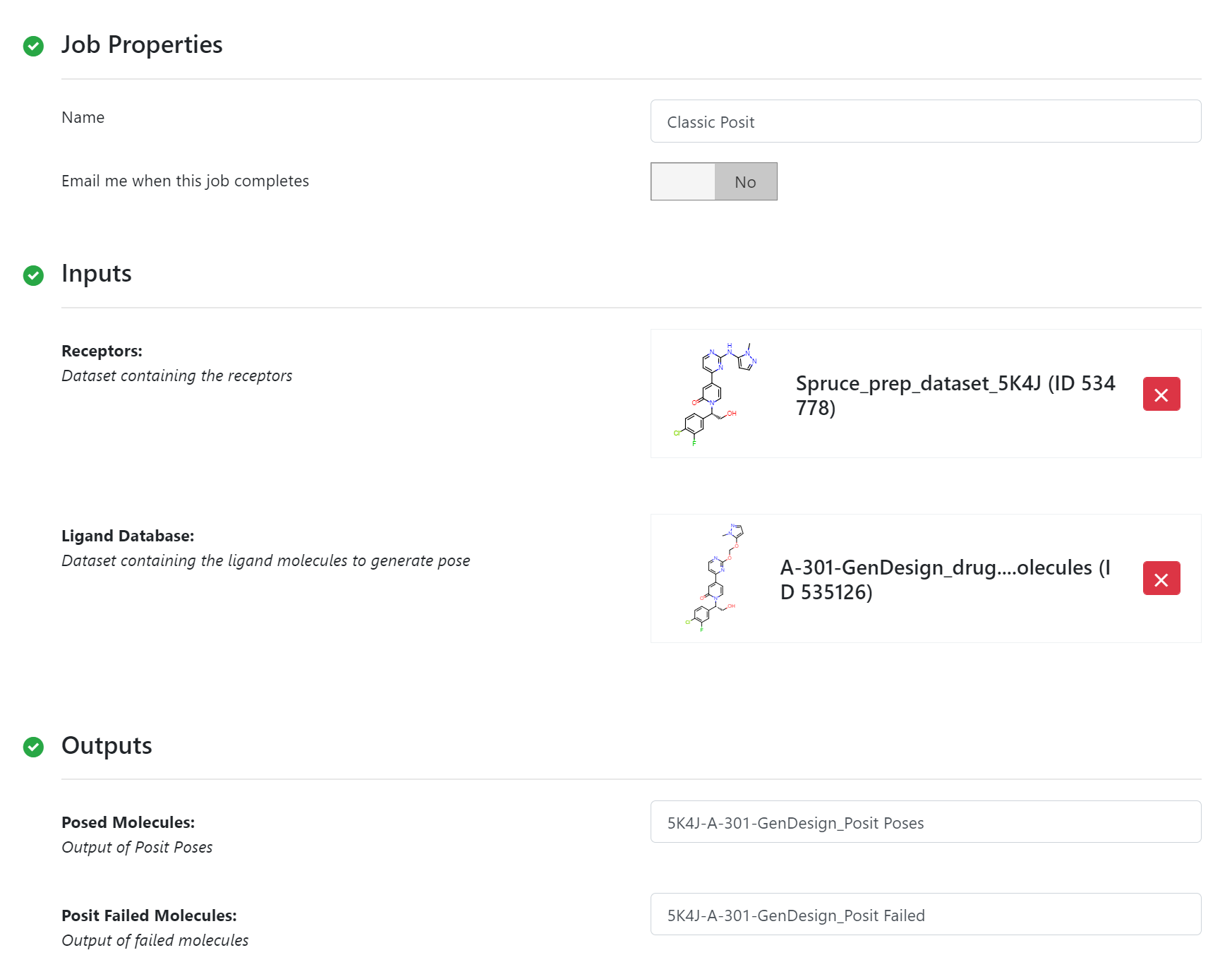
Classic Posit Floe Parameters¶
The six good poses from the Classic Posit Floe run are validated by the Short Trajectory MD with Analysis, which can be to answer questions “is this a good pose for this ligand?”, or with multiple poses, “which of these poses is best?”. There are detailed tutorials on how to use the floe. Here a brief description of using the floe on the candidates is given.
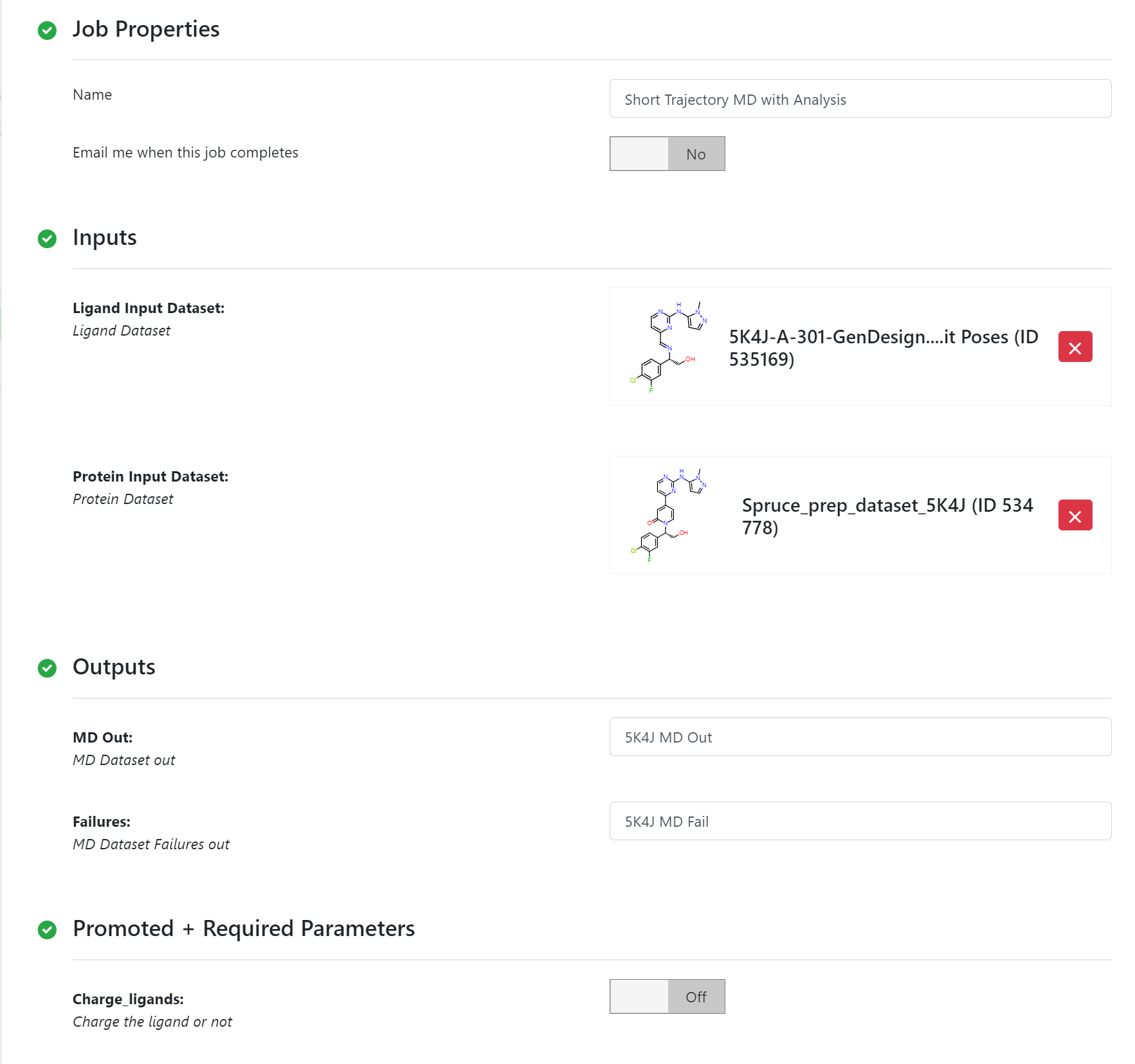
STMD Floe Parameters¶
The Charge_Lignads parameter is turned off because the candidates are already charged. There is a report to show the analysis of these poses.
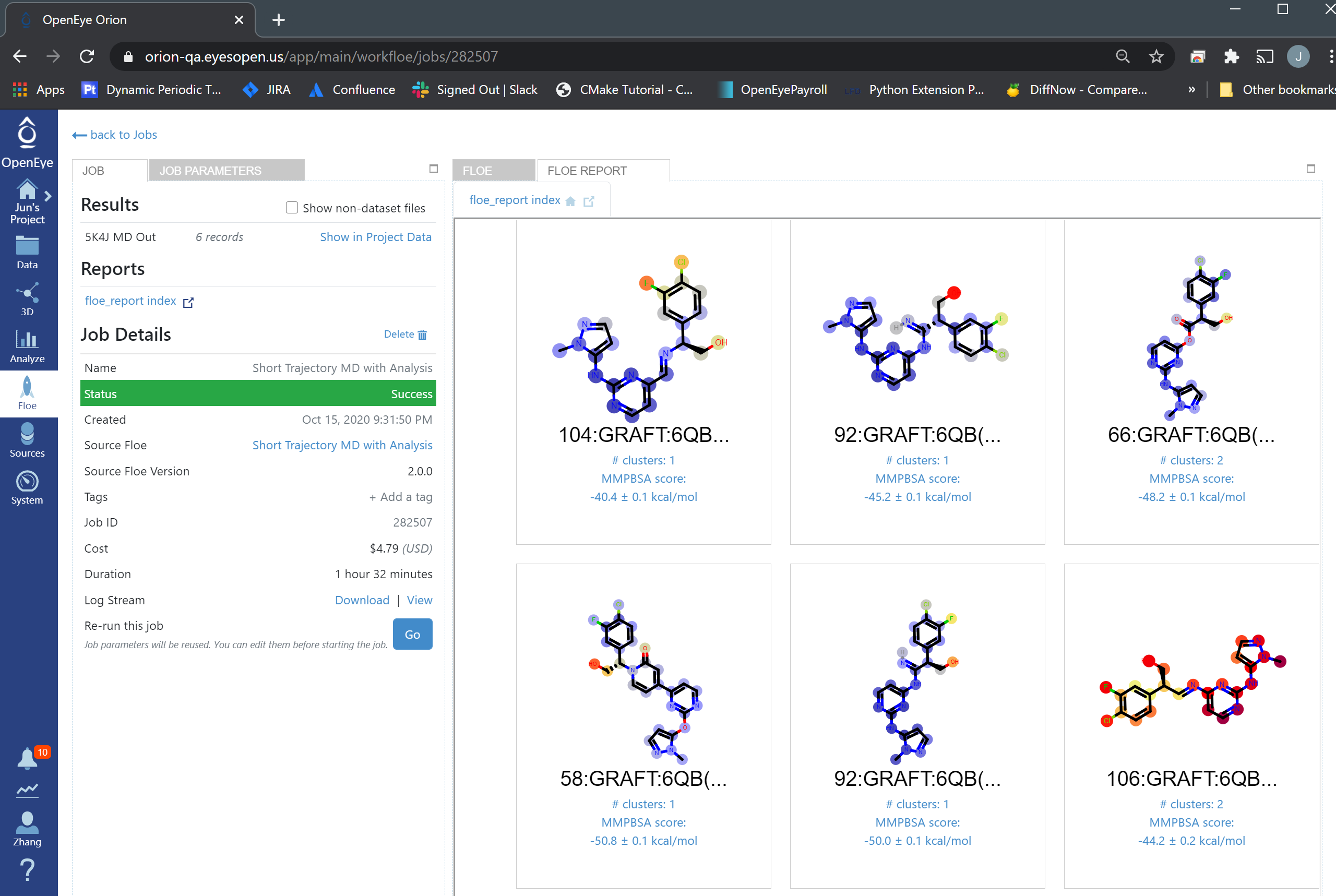
STMD Floe Report¶
And click one of them will show details of that candidate.
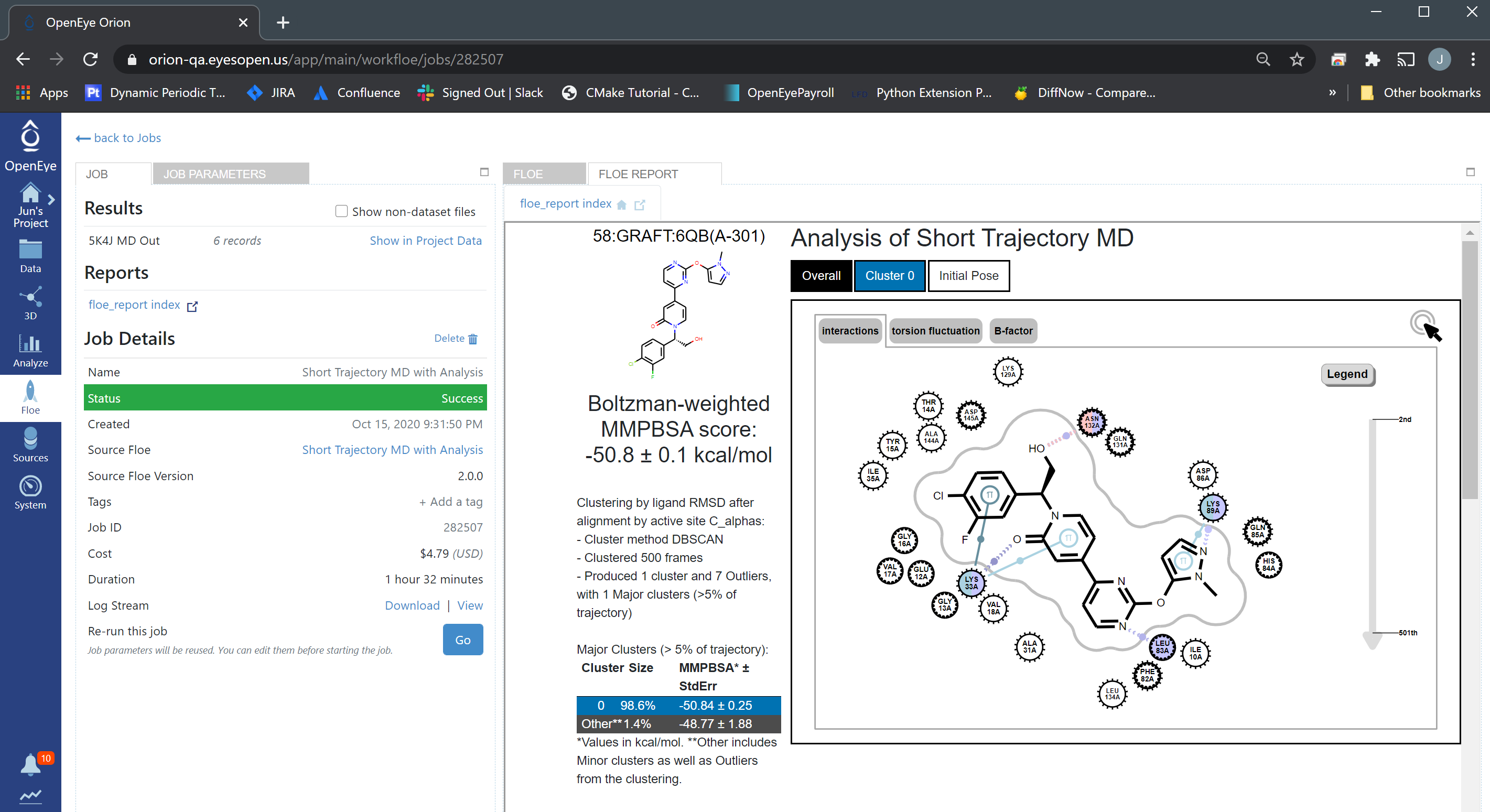
STMD Report Details¶
Selecting the output dataset in the “Data” tab and moving to the “Analyze” tab, the results for the entire dataset can be viewed at once as in the folloing figure.
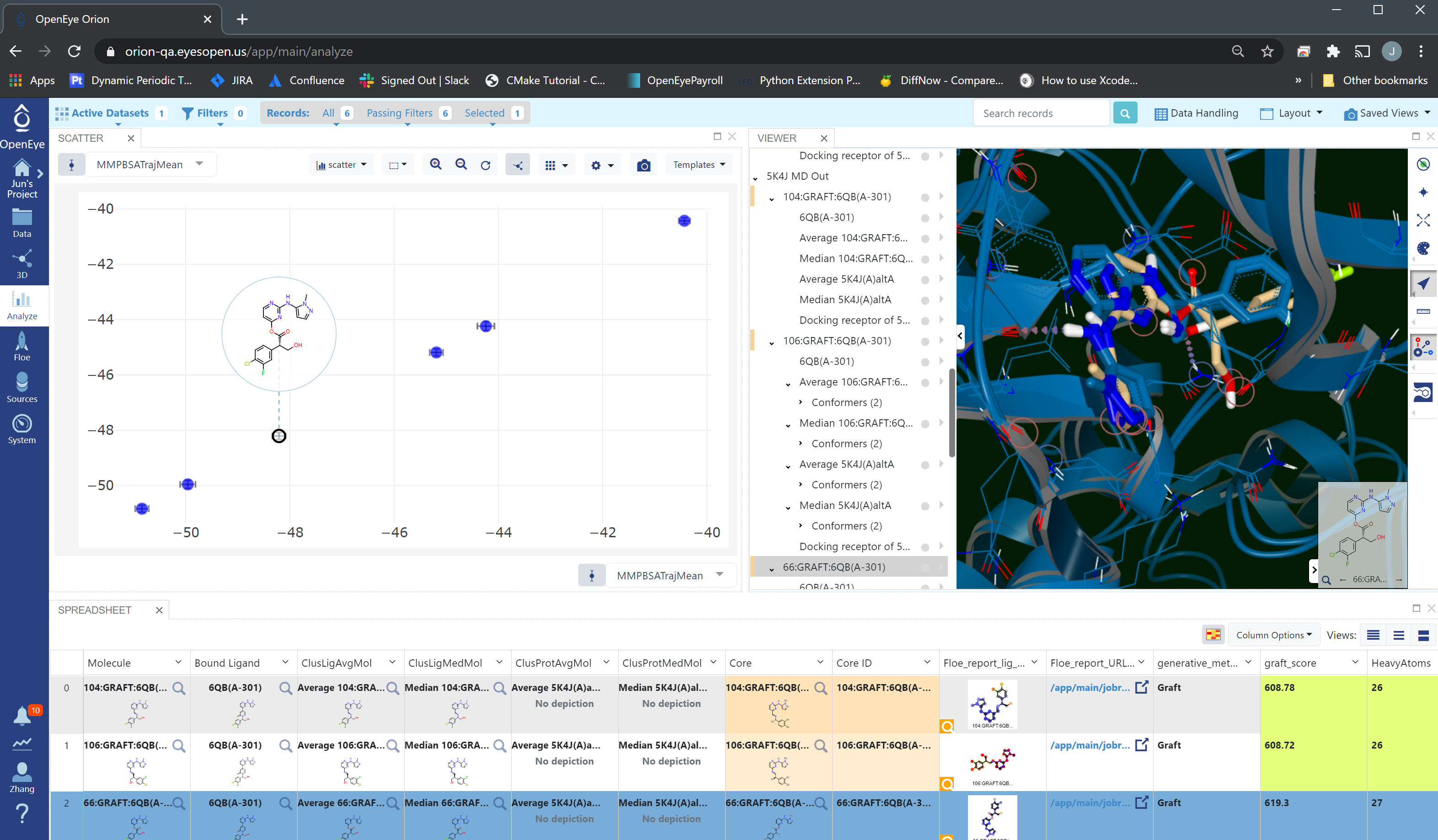
STMD Analyze Page¶
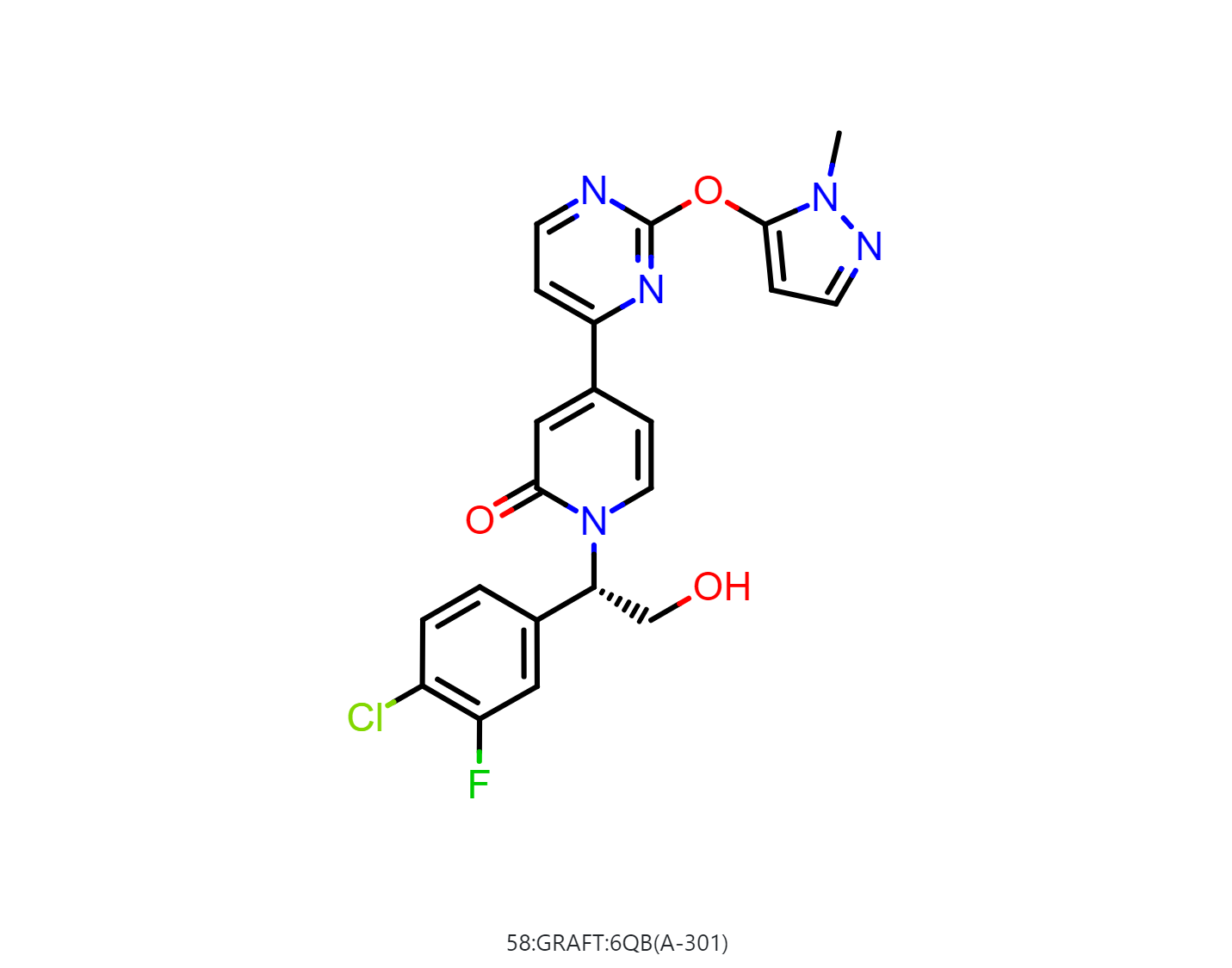
From the “Analyze” page, we can see that 58:GRAFT:6QB(A-301) has the lowest Boltzman-weighted MMPBSA score: -50.8 ± 0.1 kcal/mol, and it is the best pose and should be selected for synthesis.