MD DataRecord
MD DataRecord a brief overview
Molecular Dynamics (MD) simulations are notoriously time consuming and computational demanding. In terms of data, a lot of information need to be stored and retrieved such as atomic coordinates, atomic velocities, forces and energies just to cite only few; extracting and managing this data then becomes crucial. The MD DataRecord API simplifies the access to the MD data in the MD floe programming for Orion. In the OpenEye Datarecord model, data is exchanged between cubes in format of data records where POD data, custom objects, json data etc. are stored and retrieved by using the associated field names and types. The MD Datarecord API is built on the top of the OpenEye Datarecord, standardizing the record content data produced during MD runs in a well-structured format and providing an API point to its access.
MD DataRecord structure
The MD data produced along MD runs is structured in what is named the MD record:
the MD record contains a sub-record named MD stages where MD information is saved. This sub-record is a list of MD stage records;
each MD stage is a record itself with an associated name, type, log data, topology and MD State info. The latter is a custom object that stores data useful to restart MD runs such as, atomic positions, velocities and box information;
the MD record at the top level contains a Parmed object used to carry the whole system parametrization data
the MD record at the top level can also contain other data such as the MDComponents object used to carry info related to the different MD system parts such as ligand, protein, solvent, cofactors etc. or can contains the starting ligand and protein with their names, unique identifiers such as the flask id , cofactor ids etc.
The following picture shows the MD record structure and its main components
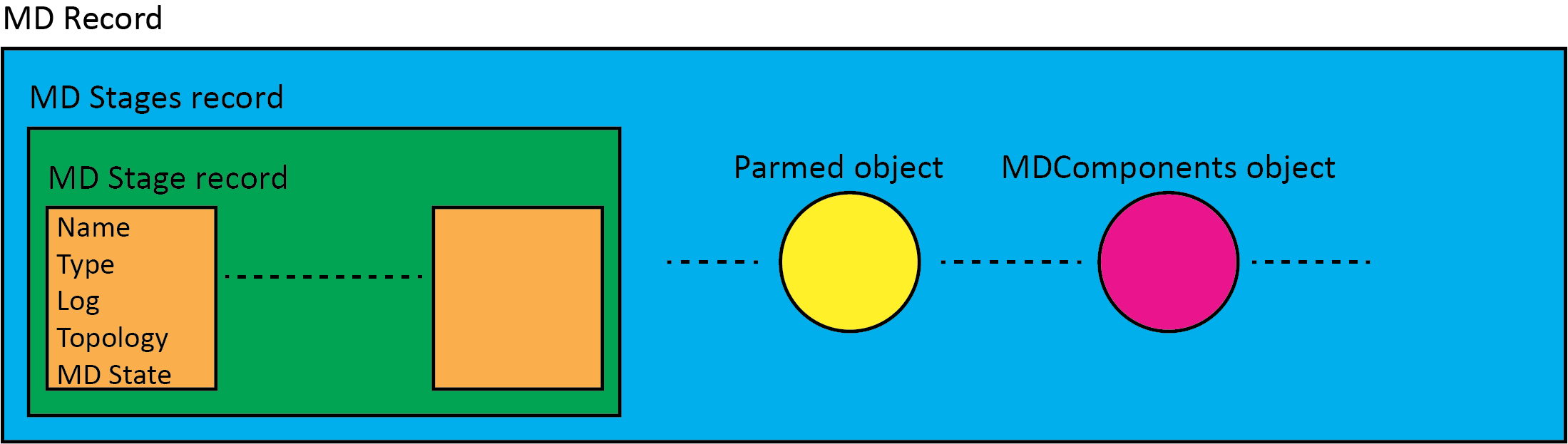
Structure of the MD Record
The MD record is user accessible by using the MDDataRecord API. In order to use it, the MDOrion package must be installed. The installation of the package also requires to have access to the OpenEye Magpie repository for some dependencies. The API has been designed to transparently work locally and in Orion. To use the API the user needs to create a MDDataRecord object starting from an OERecord object, after that getter and setter functions can be used to access the MD data (see full MDDataRecord API Documentation).
Code snippets
The following code snippets give an idea on how to use the API.
Warning
In the following examples record is an OpenEye Datarecord produced running MD floes such as Solvate and Run MD or Solvate and Run Protein-Ligand MD. Starting from the orionmdcore package v2.0.0, the datasets produced from the Short Trajectory MD with analysis floe is “ligand centric” and the MDDatarecord API cannot be directly applied to the produced records. However, the API still works on the conformer (poses) ligand sub-records which are still MD records
from orionmdcore import MDDataRecord
# To use the MD Datarecord API an MDDataRecord instance is built starting from an OERecord
md_record = MDDataRecord(record)
# At this point getters and setters can be used to
# extract/set info from/to the MD record
# MD Stage names available
stage_names = md_record.get_stages_names
# Get a MD Record Stage
md_stage_production = md_record.get_stage_by_name(stage_names[2])
# Extract the logging info from a stage
info_stage = md_record.get_stage_info(stage_names[2])
# Extract the MD State from a stage
md_set_up_state = md_record.get_stage_state(stage_names[0])
# Extract the Parmed Structure from the MD record and synchronize the
# positions, velocities and box data to the selected stage state
pmd_structure = md_record.get_parmed(sync_stage_name=stage_names[2])
# Extract the OEMol system flask from the record and its title
flask = md_record.get_flask
flask_title = md_record.get_title
# Extract the trajectory file name associated with a stage. In this
# case the stage trajectory in unpacked and the trajectory name
# can be used in any MD analysis package to be loaded
trj_name = md_record.get_stage_trajectory(stage_names[2])
# Add a new Stage to the md_stages record
md_record.add_new_stage(
stage_name="New_Stage",
stage_type="NPT",
topology=flask,
mdstate=md_set_up_state,
data_fn="test.tar.gz"
)
MDDataRecord API Documentation
The following sections document the objects in the MDDataRecord API.
- class orionmdcore.mdrecord.mdrecord.MDDataRecord(record: OERecord | None = None, **kwargs)
This Class Implements the MD Datarecord API by using getter and setter functions
- add_new_stage(stage_name: str, stage_type: str, topology: OEMol, mdstate: MDState, data_fn: str, append: bool = True, log: str | None = None, info: dict | None = None, trajectory_fn: str | int | None = None, trajectory_engine: str | None = None, trajectory_orion_ui: str | None = None) bool
This method add a new MD stage to the MD stage record
Parameters
- stage_name: String
The new MD stage name
- stage_type: String
The MD stage type e.g. SETUP, MINIMIZATION etc.
- topology: OEMol
The topology
- mdstate: MDState
The new mdstate made of state positions, velocities and box vectors
- data_fn: String
The data file name is used only locally and is linked to the MD data associated with the stage. In Orion the data file name is not used
- append: Bool
If the flag is set to true the stage will be appended to the MD stages otherwise the last stage will be overwritten by the new created MD stage
- log: String or None
Log information.
- info: Python Dictionary or None
Info Dictionary of Plain Data
- trajectory_fn: String, Int or None
The trajectory name for local run or id in Orion associated with the new MD stage
- trajectory_engine: String or None
The MD engine used to generate the new MD stage. Possible names: OpenMM or Gromacs
- trajectory_orion_ui: String
The trajectory string name to be displayed in the Orion UI
Returns
- boolean: Bool
True if the MD stage creation was successful
- create_collection(name: str, v2: bool = True) bool
This method sets a collection field on the record to be used in Orion
Parameters
- namestr
A string used to identify in the Orion UI the collection.
- sessionOrionSession
An Orion Session object.
- v2bool, default True
Whether to create a v2 collection.
Returns
- booleanbool
True if the collection creation in Orion was successful otherwise False
- property delete_parmed: bool
This method deletes the Parmed object from the record. True is returned if the deletion was successful.
Parameters
- session: Orion Session
An Orion Session object
Returns
- booleanBool
True if Parmed object deletion was successful
- delete_stage(stage_id: int | str = -1) bool
This method deletes an MD stage selected by passing its index or name. If the stage cannot be found an exception is raised.
Parameters
- idx: int, str
The MD stage index or name
Returns
- boolean: bool
True if the deletion was successful
- delete_stage_by_idx(idx: int) bool
This method deletes an MD stage selected by passing its index. If the stage index cannot be found an exception is raised.
Parameters
- idx: int
The MD stage index
Returns
- boolean: bool
True if the deletion was successful
- delete_stage_by_name(name: str = 'last') bool
This method deletes an MD stage selected by passing the string name. If the string “last” is passed (default) the last MD stage is deleted. If no stage name has been found an exception is raised.
Parameters
- name: str
The MD stage name
Returns
- boolean: bool
True if the deletion was successful
- property delete_stages: bool
This method deletes all the record stages
Parameters
Returns
- booleanBool
True if the deletion was successful
- property get_conf_id: int
This method returns the identification field CONF ID present on the record
Parameters
Returns
- conformed_id: Int
The conformer id
- property get_extra_data_tar: str
This method returns the directory file name where extra data file tar has been unpacked
Parameters
Returns
- directory file name: String
The directory file name
- property get_flask: OEMol
This method returns the flask molecule present on the record
Parameters
Returns
- flask: OEMol
The flask present on the record otherwise an error is raised
- property get_flask_id: int
This method returns the integer value of the flask identification field present on the record
Parameters
Returns
- flask_idInt
The unique flask identifier
- property get_last_stage: MDDataRecord
This method returns the last MD stage of the MD record stages
Parameters
Returns
- record: MDDataRecord
The last stage of the MD record stages
- property get_lig_id: int
This method returns the ligand identification field present on the record
Parameters
Returns
- ligand_id: Int
The ligand identification id number
- property get_ligand: OEMol
This method returns the ligand molecule present on the record
Parameters
Returns
- ligand: OEMol
The ligand molecule if the ligand has been set on the record otherwise an error is raised
- property get_ligand_traj: OEMol
This method returns the ligand molecule where conformers have been set as trajectory frames
Returns
- multi_conformer_ligand: OEMol
The multi conformer ligand
- property get_md_components: MDComponents
This method returns the MD Components if present on the record
Parameters
Returns
- md_components: MDComponents
The MD Components object
- property get_primary: OEMol
This method returns the primary molecule present on the record
Parameters
Returns
- record: OEMol
The Primary Molecule
- property get_protein: OEMol
This method returns the protein molecule present on the record
Parameters
Returns
- proteinOEMol
The protein molecule if the protein has been set on the record otherwise an error is raised
- property get_protein_traj
This method returns the protein molecule where conformers have been set as trajectory frames
Parameters
- session: Orion Session
An Orion Session obj
Returns
- multi_conformer_protein: OEMol
The multi conformer protein
- property get_record: OERecord
This method returns the record.
Parameters
Returns
record: OERecord
The record to be passed with the cubes
- get_stage(stage_id: str | int = -1) MDDataRecord
This method returns a MD stage selected by passing an index or name. If the stage is not found an exception is raised.
Parameters
- idx: int, str
The index or the name of the stage to retrieve
Returns
- record: MDDataRecord
The selected MD stage
- get_stage_by_idx(idx: int) MDDataRecord
This method returns a MD stage selected by passing an index. If the stage is not found an exception is raised.
Parameters
- idx: int
The stage index to retrieve
Returns
- record: MDDataRecord
The MD stage selected by its index
- get_stage_by_name(name: str = 'last') MDDataRecord
This method returns a MD stage selected by passing the string stage name. If the string “last” is passed (default) the last MD stage is returned. If multiple stages have the same name the first occurrence is returned. If no stage name has been found an exception is raised.
Parameters
- name: str
The MD stage name
Returns
- record: MDDataRecord
The MD stage selected by its name
- get_stage_info(stage_id: int | str = -1) str
This method returns the info related to the selected stage name. If no stage name is passed the last stage is selected.
Parameters
- name: str
The MD stage name
Returns
- info_string: str
The info associated with the selected MD stage otherwise None
- get_stage_logs(stage_id: int | str = -1) str
This method returns the logs related to the selected stage name. If no stage name is passed the last stage is selected.
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- info_string: str
The info associated with the selected MD stage
- get_stage_state(stage_id: str | int = -1) MDState
This method returns the MD State of the selected stage name. If no stage name is passed the last stage is selected
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- stateMDState
The MD state of the selected MD stage
- get_stage_topology(stage_id: str | int = -1) OEMol
This method returns the MD topology of the selected stage name. If no stage name is passed the last stage is selected.
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- topologyOEMol
The topology of the selected MD stage
- get_stage_trajectory(stage_id: str | int = -1) str | None
This method returns the trajectory file name associated with the md data. If the trajectory is not found None is return
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- trajectory_filename: str, None
Trajectory file name if the process was successful otherwise None
- property get_stages: list[MDDataRecord]
This method returns the MD stage record list with all the MD stages.
Parameters
Returns
- record_list: list
The MD stages record list
- property get_stages_names: list[str]
This method returns the list names of the MD stages.
Parameters
Returns
- list: list
The MD stage name list
- property get_title: str
This method returns the title present on the record
Parameters
Returns
- titleString
The title string if present on the record otherwise an error is raised
- property get_water_traj: OEMol
This method returns the water molecule where conformers have been set as trajectory frames
Parameters
Returns
- multi_conformer_water: OEMol
The multi conformer water
- property has_conf_id: bool
This method checks if the identification field CONF ID is present on the record
Parameters
Returns
- booleanBool
True if the conformer id field is present on the record otherwise False
- property has_extra_data_tar: bool
This method returns True if extra data file in tar format is attached to the record
Return
- boolean: Bool
True if extra data file is present on the record otherwise False
- property has_flask_id: bool
This method checks if the flask identification field is present on the record
Parameters
Returns
- boolean: Bool
True if the flask ID field is present on the record otherwise False
- property has_lig_id: bool
This method checks if the ligand identification field is present on the record
Parameters
Returns
- booleanBool
True if the ligand identification field is present on the record otherwise False
- property has_ligand: bool
This method returns True if ligand molecule is present on the record
Parameters
Returns
- booleanBool
True if the ligand molecule is present on the record otherwise False
- property has_ligand_traj: bool
This method checks if the multi conformer ligand is on the record.
Parameters
Returns
- booleanBool
True if the multi conformer ligand is on the record otherwise False
- property has_md_components: bool
This method returns True if the MD Components object is present on the record
Return
- boolean: Bool
True if the md components object is present on the record otherwise False
- property has_parmed: bool
This method checks if the Parmed object is on the record.
Parameters
Returns
- booleanBool
True if the Parmed object is on the record otherwise False
- property has_protein: bool
This method returns true if the protein molecule is present on the record
Parameters
Returns
- booleanBool
True if the protein molecule is present on the record otherwise False
- property has_protein_traj: bool
This method checks if the multi conformer protein is on the record.
Parameters
Returns
- booleanBool
True if the multi conformer protein is on the record otherwise False
- has_stage(stage_id: int | str = -1) bool
This method returns True if MD stage selected by passing the string name is present on the MD stage record otherwise False.
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- boolean: bool
True if the MD stage name is present on the MD stages record otherwise False
- has_stage_info(stage_id: int | str = -1) bool
This method returns True if MD stage selected by passing the string name has infos present on the MD stage record otherwise False.
Parameters
- stage_id: str, int
The MD stage name or index
Returns
- boolean: bool
True if the MD stage has info otherwise False
- has_stage_name(name: str) bool
This method returns True if MD stage selected by passing the string name is present on the MD stage record otherwise False.
Parameters
- name: str
The MD stage name
Returns
- boolean: bool
True if the MD stage name is present on the MD stages record otherwise False
- property has_stages: bool
This method returns True if the record has a MD record list otherwise False
Parameters
Returns
- booleanBool
True if the record has a list of MD stages otherwise False
- property has_title: bool
This method checks if the Title field is present on the record
Parameters
Returns
- booleanBool
True if the Title field is resent on the record otherwise False
- property has_water_traj: bool
This method checks if the multi conformer water is on the record.
Parameters
Returns
- booleanBool
True if the multi conformer water is on the record otherwise False
- set_conf_id(conf_id: int) bool
This method sets the identification field for the conformer on the record
Parameters
- conf_id: Int
An identification integer for the record
Returns
- booleanBool
True if the conformed id has been set on the record otherwise an error is raised
- set_extra_data_tar(tar_fn: str, shard_name: str = '') bool
This method sets the extra data file on the record
Parameters
- tar_fn: String
The compressed data file name
- shard_name: String
In Orion tha shard will be named by using the shard_name
Returns
- boolean: Bool
True if the setting was successful
- set_flask(flask: OEMol) bool
This method sets the flask molecule on the record
Parameters
- flaskOEMol
The flask molecule to set on the record
Returns
- record: Bool
True if the flask molecule has been set on the record otherwise an error is raised
- set_flask_id(id: int) bool
This method sets the integer value of the flask identification field on the record
Parameters
- id: Int
An integer value for the flask identification field
Returns
- booleanBool
True if the flask identification ID has been set as an integer on the record
- set_lig_id(sys_id: int) bool
This method sets the ligand identification field on the record
Parameters
- sys_id: Int
An integer value for the ligand identification field
Returns
- boolean: Bool
True if the value for the ligand identification field was successfully set on the record
- set_ligand(ligand: OEMol) bool
This method sets the ligand molecule on the record
Parameters
Returns
- boolean: Bool
returns True if the ligand has been set on the record otherwise an error is raised
- set_ligand_traj(ligand_conf: OEMol, shard_name: str = '') bool
This method sets the multi conformer ligand trajectory on the record
Parameters
- ligand_conf: OEChem
Th multi conformer ligand trajectory
- shard_name: String
In Orion tha shard will be named by using the shard_name
Returns
- boolean: Bool
True if the setting was successful
- set_md_components(md_components: MDComponents) bool
This method sets the MD Components on the record
Parameters
- md_components: MDComponents
The MD Components instance
Returns
- booleanBool
True if the md components field was successfully set on the record
- set_parmed(pmd: Structure, sync_stage_name: str | None = None, shard_name: str = 'Parmed') bool
This method sets the Parmed object. Return True if the setting was successful. If sync_stage_name is not None the parmed structure positions, velocities and box vectors will be synchronized with the MD State selected by passing the MD stage name
Parameters
- pmd: Parmed Structure object
The Parmed Structure object to be set on the record
- sync_stage_name: String or None
The stage name that is used to synchronize the Parmed structure
- shard_name: String
In Orion tha shard will be named by using the shard_name
- session: Orion Session
An Orion Session obj
Returns
- booleanBool
True if the setting was successful
- set_primary(primary_mol: OEMol) bool
This method sets the primary molecule on the record
Parameters
- primary_mol: OEMol
The primary molecule to set on the record
Returns
- boolean: Bool
True if the primary molecule has been set on the record
- set_protein(protein: OEMol) bool
This method sets the protein molecule on the record
Parameters
Returns
- boolean: Bool
returns True if the protein has been set on the record otherwise an error is raised
- set_protein_traj(protein_conf: OEMol, shard_name: str = '') bool
This method sets the multi conformer protein trajectory on the record
Parameters
- protein_conf: OEMol
The multi conformer protein trajectory
- shard_name: String
In Orion the shard will be named by using the shard_name
Returns
- boolean: Bool
True if the setting was successful
- set_stage_info(info_dic: dict, stage_id: int | str = -1)
This method sets the stage info field on the selected stage by name
Parameters
- stage_id: str, int
The MD Stage name or index
- info_dic: dict
The dictionary containing the Plain Data info to save
Returns
- booleanbool
True if the system Title has been set on the record
- set_title(title: str) bool
This method sets the system Title field on the record
Parameters
- title: String
A string used to identify the molecular system
Returns
- booleanBool
True if the system Title has been set on the record
- set_water_traj(water_conf: OEMol, shard_name: str = '') bool
This method sets the multi conformer water trajectory on the record
Parameters
- water_conf: OEChem
Th multi conformer water trajectory
- shard_name: String
In Orion the shard will be named by using the shard_name
Returns
- boolean: Bool
True if the setting was successful
- class orionmdcore.mdrecord.mdrecord.MDStageRecord(record: OERecord | None = None, **kwargs)
This Class is used to store for and extract data from the MD stages.