Release Notes
4.1.10 November 2024
Bugfixes
This version addressed a rare issue in the Gigadock Warp floe where the GPU memory could become fragmented while training the AI score model, causing the floe to fail. The updated floe will now complete normally in this circumstance and not fail.
4.1.9 October 2024
Bugfixes
The Gigadock Warp floe no longer attempts to use Cadence data center GPUs by default.
4.1.7 July 2024
Highlights
CXSMILES support has been added to the Prepare Giga Collections Floe.
Bug Fixes
A bug has been fixed in the Gigadock Warp Floe that could cause the Floe to hang for a long time and then fail if AWS S3 is under load and transferring data slower than expected.
The Options: Model Training –> Fraction Train parameter for the Gigadock Warp Floe can now be set as low as 0.001 as stated in the parameter description. A bug in the prior release prevented the value from being set below the default of 0.01.
A bug has been fixed in the Gigadock, Gigadock Warp, and FastROCS Plus Floes from including individual dock score components for each molecule in the output when these values are requested by the user. They are only expected to be in the output if the user specifies the parameter output fields for these components; these parameters are in the Output Fields parameter category for all three floes.
A typo has been fixed in the tutorial documentation to correctly indicate that the FastROCS Plus Floe produces seven output hit lists. It had incorrectly indicated five.
4.1.5 February 2024
Highlights
The Gigadock Warp Floe has been upgraded with better featurization of the molecules and now uses a neural net model rather than a linear model when docking more than ~100M molecules. Output hit lists from Gigadock Warp should now be expected to even more closely match the hit list that would be created by running the Gigadock Floe and cost 30%-40% less.
Features
Gigadock Warp now uses a neural net rather than a linear score model when docking more than 93,460,000 molecules. Below this threshold, it still uses a linear model.
Gigadock Warp now uses a 4K Tree fingerprint, FastROCS Shape and Combo Tanimotos, and simple 2D properties to featurize the molecules. The previous version used MACSS166 fingerprints in place of the 4K Tree fingerprint.
Changes
The “Predicted Score” field has been removed from the Gigadock Warp output.
Gigadock Warp now always uses AWS spot instance for all major parallel cubes that don’t use a GPU.
Gigadock Warp now defaults to docking 4% of the molecule in the final docking stage (was 8%).
The following parameters have been added to the Gigadock Warp Floe:
Options: Model Training -> Fraction Train
Options: Model Training -> Final Dock Fraction
Options: Hardware -> FastROCS Instance Types
Options: Hardware -> FastROCS Spot Policy
Bugfixes
A bug was fixed where Gigadock Warp would fail to output a hit list if the Options: Output Fields -> Docked Pose Field was specified.
An issue was fixed where the Collection Info Floe could fail when processing a 10B-sized Gigadock collection.
An issue was fixed where Collection Info could have negative counts in the report’s histograms if the collection was very large (~>5B).
4.0.2 September 2023 (Large Scale Floes Hotfix)
Changes
Filter Collection can now take multiple input collections of the same type (e.g., FastROCS or Gigadock)
Bugfixes
Fixed issue with Gigadock Warp hanging when processing extremely large collections.
Fixed failure in FastROCS Plus if ROCS re-scoring is turned off and dock re-scoring is not turned on.
Fixed issue with ‘Prepare Giga Collection’ intermittently failing in the beginning of the floe if given large input file.
Fixed missing Gigadock Warp html documentation
4.0.0 July 2023 (Orion Floe 2023.1 Release)
Highlights
The Gigadock Warp Floe has been upgraded to use an AI model and will now cost less than the previous version and produce hit lists more similar to the hit list the Gigadock Floe would produce given the same input. See Gigadock Warp Details for more details.
Changes
The parameter Options: Re-scoring -> Number of Molecules to Re-score can now be set up to 100 M (the previous limit was 10M).
Gigadock Warp now requires 1 M or more input molecules to dock in order to ensure it has enough training data to build an AI model of the score.
The following parameters have been removed from the Gigadock Warp Floe as part of the internal reworking of Gigadock Warp to now support AI learning of the docking scores.
Options: Advanced -> Random Dock Fraction
Options: Advanced -> Final Dock Fraction
Options: Advanced -> Number of FastROCS Queries
Options: Advanced -> Cluster FastROCS Queries
Bugfixes
An issue was fixed in FastROCS Plus where the consensus hit lists incorrectly had an extra field named “Pareto Dominance Rank,” in addition to a expected field “Pareto Rank.”
3.4.5 December 2022 (Orion 2022.4 Release)
New Features
FastROCS Plus now supports any type of Shape Query in the ROCS Re-scoring step. The core FastROCS screen is still restricted to simpler Shape Queries.
FastROCS Plus now supports searching by shape only. I.e, Shape Tanimoto, Ref Shape Tversky and Fit Shape Tversky are now additionally supported as similarity types.
The Filter Collection and Prepare Giga Collections floes now create a floe report describing the number of molecules that pass each filtering step.
Bugfixes
The Filter Collection and Prepare Giga Collections floes should now halt with an error when the specified filters eliminate all molecules.
3.3.4 September 2022
Bugfixes
Collection based cubes now validate that downloaded shard data is the correct size, and retry the download if it is not. This protects against an extremely remote possibility that shard reads could lose data or hang the floe.
Cluster poses now properly clusters poses when ‘Options -> Single Conformer/Pose Input’ is switched from its default value of ‘On’ to ‘Off’.
The parameter ‘Options: Advanced -> Final Dock Fraction’ in the Gigadock Warp can no longer be set higher than 0.1. (There was an accidental regression in 3.3.0.) The default value of 0.08 is unchanged.
3.3.0 June 2022 (Orion 2022.2 release)
Features
New Floe : Multi Query 2D Similarity.
FastROCS Plus now automatically outputs two additional datasets by default, ‘FastROCS Novelty Hit List’ and ‘ROCS Novelty Hit List’. These hit lists contain molecules from the FastROCS and the ROCS Re-scoring computations respectively that tend to have high 3D and low 2D similarity.
The Filter Collection and Prepare Giga Collections floes can now optionally take a set of known molecules and then filter the collection molecules based on their 2D similarity to the known molecules (see the ‘Options: Known Molecules’ parameter group in both these floes).
The Prepare Giga Collections now optionally accepts a dataset as input.
FastROCS Plus output hit lists now includes a field with the 2D Tanimoto similarity to the query.
Changes
Filter Collection and Prepare Giga Collections now applies filtering from Options -> Keep this Fraction before all other filters, rather than after.
Bugfixes
Prepare Giga Collections no longer losses ~0.07% of molecules from the Gigadock collection when preparing large collections (the FastROCS Collection was unaffected by this bug).
Prepare Giga Collections no long creates an empty Gigadock collection if fewer than 1000 molecules are prepared.
Filter Collection and Prepare Giga Collections no longer filter out all molecules if both a custom and builtin OEFilter are used.
Gigadock Warp output molecules now have explicit rather than implicit hydrogens.
Prepare Giga Collections will now fail if run with no input.
Batch FastROCS and FastROCS Plus output query datasets now have molecule queries as the primary molecule.
Fixed an out or memory issues in the FastROCS Plus floe that could occur in rare circumstances.
3.2.0 April 2022 (Orion 2022.1 release)
Changes
The Prepare Giga Collections and Batch FastROCS floes now by default can use a wider variety of GPU instances This should help reduce overall runtime of the floes.
Minor Features
The Prepare Giga Collections floe now by default adds a field “Enantiomer Title” to the output collections that contains the title of the molecule with a postfix index to identify enantiomers of input molecule with unspecified stereo that were stereochemically enumerated.
The FastROCS Plus floe now has an option to output a collection of up to 10M of the top scoring FastROCS molecules.
Bugfixes
Fixed a bug where the FastROCS Plus floe could fail due to an out of memory error if Options: Advanced: Number of Molecules to Re-score is set to a value much higher than the default.
Fixed an issue where Prepare Giga Collections could hang and the end of the floe waiting for the docking collection to close. This issue only affected preparation of large collections (e.g., Billions) and when using CPU rather than GPU Omega.
Fixed an issue in FastROCS Plus that caused more than one docked pose per molecule to be used when ‘Options: Advanced -> Query Conformer Generation Mode’ was set to ‘dock’.
Fixes several grammatical error in the FastROCS Plus and Batch FastROCS floe and parameter descriptions.
Fixed an issue in the documentation of floe in the large scale floes package where the python names of parameters were not correct. (The python name of the parameter is the name used when launching a job with ocli).
3.1.7 Dec 2021 (Orion 2021.2.1 release)
Highlights
This release contains a new floe Gigadock Warp that approximates a full Gigadock run using a combination of Docking and FastROCS. Gigadock Warp is ~8-10x less costly to run than a full Gigadock job and when docking billions of molecule recovers 70% of the same molecules the full Gigadock job does in the top 10K hit list.
See also
For a tutorial on running Gigadock Warp see Dock Ten Million Molecules with Gigadock Warp and Analysis with Freeform Consensus
For a explanation of how Gigadock Warp works see Gigadock Warp Details.
General Notice
GPU instances on Amazon Web Services (AWS) have been in high demand recently. This can result in long run times for floes that uses GPUs, i.e., FastROCS Plus, Batch FastROCS, Prepare Giga Collections and Gigadock Warp. To help reduce the chance of encountering this issue these floes have been modified to by default use older AWS GPU instances that are generally more available but slight less cost efficient (~25% more GPU cost). These floes now expose parameters to allow specifying more the more efficient, but sometimes less available instances when a floe is run. See the Changes section of these release notes for details on individual floes.
New Features
New Floe : Gigadock Warp approximates a full Gigadock run using a combination of Docking and FastROCS.
See also
For a tutorial on running Gigadock Warp see Dock Ten Million Molecules with Gigadock Warp and Analysis with Freeform Consensus
For a explanation of how Gigadock Warp works see Gigadock Warp Details.
New Floe : Pareto Frontier Consensus finds the best records in an input dataset based on two or more numeric values in the dataset using a Pareto Frontier analysis.
See also
The Batch FastROCS floe now by default pre-appends each output datasets with the name of input query it is associated with.
The Batch FastROCS floe now has an additional output dataset for each query containing the cluster heads of the output hit list. Full clustering information is still present in the primary hit lists.
Changes
The FastROCS Plus no longer outputs the Similarity Combo, Color Similarity and Shape Similarity fields. These fields were duplicates of the output fields Tanimoto Combo, Color Tanimoto, and Shape Tanimoto (or fields with Tversky instead of Tanimoto if Tversky scoring is used) which were and continue to be outputted.
The GigaDock floe has been renamed the Gigadock floe.
The Giga Docking Collection to Hi-res FastROCS Collection has been renamed the Gigadock Collection to Hi-res FastROCS Collection floe.
The FastROCS Plus (Keywords: Shape, Docking, Consensus, Collection, Virtual Screening) floe has been renamed the FastROCS Plus floe.
The Batch FastROCS (Keywords: Shape, Collection, Virtual Screening) floe has been renamed the Batch FastROCS floe.
The FastROCS Plus floe now uses AWS g3 GPU instance by default (previous it used g4dn.2xlarge). Parameters for choosing the AWS GPU instance to use are now exposed (see the GPU Hardware parameter group).
The Batch FastROCS floe now uses AWS g3 GPU instance by default (previous it used g4dn.xlarge). Parameters for choosing the AWS GPU instance to use are now exposed (see the GPU Hardware parameter group).
The Prepare Giga Collections now uses CPU Omega by default (previously it used GPU Omega). This can be adjusted with the Conformer Generation Settings -> Use GPU Omega setting.
When processing a tab or comma separated file Prepare Giga Collections will now, by default, name the field on the output giga docking collection ‘Molecule’ rather than using the column title from the csv/tsv file (see the new Format Specific Settings -> Molecule Field Name For TSV/CSV parameter). This change was made because in practice the column is often named ‘SMILES’ and this field name propagates to the docked pose field of the output hit lists of the Gigadock floe, where ‘SMILES’ is not an appropriate name.
Fixes
Fixed a bug in the Batch FastROCS floe that causes it not to write out the fields with the standard names for ROCS scores, i.e., Tanimoto Combo, Color Tanimoto, and Shape Tanimto or Tversky Combo, Color Tversky, and Shape Tvserky when Tversky similarity is selected. The floe was previously writing out fields with Similarity in place of Tanimoto or Tversky in the field names.
In the FastROCS Plus floe, fixed the description of the parameter Options: Advances -> Query Conformer Generation Modes which incorrectly referred to ‘freeform’ as an available conformer generation mode.
Fixed a bug in the FastROCS Plus floe that caused it to ignore all design units supplied to the Inputs -> Design Unit(S) (Optional) parameter if Options: Advanced -> ROCS Re-Scoring Mode was set to ‘Off’.
Fixed a bug in Filter Collection that causes it to lose compression on the molecules of FastROCS collections. In practice this resulted in a functional but slightly larger than necessary output collection when processing FastROCS collections (these collections were not functional in the older deprecated Multi-Query Ligand-Based Virtual Screening with FastROCS and SubROCS floe).
Fixed a bug in the Collection Info, Prepare Giga Collections and Filter Collection floes that causes them to use more bandwidth than necessary reading collections.
Suppressed rare XlogP calculation warning for individual atoms when running Prepare Giga Collections and Filter Collection. While rare, when processing billions of molecules they tended to flood the log with thousands of un-actionable warnings.
3.0.3 Oct 2021 (hotfix)
Fixes
Fixed a bug that could cause docking to fail when running in multi-receptor Hybrid mode.
Removed a development parameter that was accidentally exposed in the 3.0.2 release.
3.0.2 Jun 2021 (Orion 2021.1 release)
General Notice
The functionality of the “Multi-Query Ligand-Based Virtual Screening with FastROCS and SubROCS” from the “OpenEye Ligand-Based Virtual Screening” package has been absorbed into the Batch FastROCS in this package.
New Features
All floes now have much improved parameter organization in the floe launch UI.
New Floes
FastROCS Plus FastROCS search with optional re-scoring and consensus scoring of best fastROCS molecules with Docking and ROCS.
Batch FastROCS FastROCS search with ROCS re-score of best FastROCS molecules. A separate hit-list is produce for each query.
FreeForm Pose Calculates the Freeform Delta G of docked poses with optional Delta G filtering of poses.
Cluster Poses Clusters docked poses, or alternatively ROCS overlays, based on the 3D similarity.
Sample Collection Converts a random sample of a FastROCS or GigaDocking collection into a Dataset.
Gigadock Collection to Hi-res FastROCS Collection Converts a Giga Docking Collection into a fastROCS collection with a max of 200 conformers per molecule, as opposed to a max of 10 conformers per molecule in a standard fastROCS collection.
Gigadock now supports multiple receptors/design units.
Prepare Giga Collections now supports GPU Omega (enabled by default) which can reduce floe cost by up to 40%.
Prepare Giga Collections now retains the original SMILES string when processing SMILES files or command/tab separated files containing SMILES.
Minor Features
Prepare Giga Collections now has a option to accept the input tautomer state rather than setting an appropriate tautomer state.
Collection Info can now accept files or datasets as input in addition to collections.
Fixes
Fixed issue where floes could not read collections created by very old versions of Orion.
2.0.6 April 2021 (hotfix)
Fixes
Fixed issue where a Gigadock job could fail if the Orion stack is under very heavy load.
Fixed issue where Prepare Giga Collections ignored the setting of the ‘Mol Title Field’ in some circumstances.
2.0.0 Nov 2020 (Orion 2020.3 release)
General Notice
Parameters for setting the output fields for the docked molecule and score have been temporarily removed along with the parameter specifying the input molecule field. These parameters are very rarely needed and have been set by users not fully understanding their impact. These parameters are planned to return in a future release once the FLOE UI has been improved to provide better clarity on when to set them.
New Features
No new feature in this release.
Fixes
Added duplicate checking to ‘Prepare Giga Collections’. Only applied to sequential molecules.
Fixed issue with molecules from a dataset (not collection) being docking by ‘Giga Dock’ floe being lost rather than being sent to the Restart Collection if the cost threshold was exceeded.
Fixes issue with ‘Prepare Giga Collections’ not deleting its temporary collection.
Better ordering of parameters (better categorization is coming in a future release).
Improved the efficiency of the Giga-Dock floe by about 15%
Protected the giga-docking floe against the unlikely case of 12h timeout docking an individual shard (a full docking collection consists of ~1M shards) of molecule to a gigantic active site (>~2000 cubic Angstroms).
0.1.0 August 2020 (Orion 2020.2 release)
General Notice
Initial release of Large Scale Floes package.
This package include the functionality of the OpenEye Giga Docking Floes and OpenEye Scientific Floes packages, both of which have been deprecated. (Exception: The torsion scanning floes from OpenEye Scientific Floes were moved into the OpenEye QM Floes package).
The Giga Dock (FRED) and Giga Dock (HYBRID) floes from OpenEye Giga Docking Floes have been merged into a single floe Giga Docking which has a parameter to select the docking mode (i.e., FRED or HYBRID).
The Step 1,2 & 3 collection preparation floes from OpenEye Scientific Floes have been merged into a single floe Prepare Giga Collections floe.
New Features
The collection preparation floe filters are now significantly more configurable. Some of the new optional filtering options are:
Filtering by SMARTS pattern (either exclude or require)
Filtering with a custom OEFilter file
Random filtering. This allows the creating of collection that contain a random subset of the input molecule for testing purposes.
The collection preparation floe now accepts a wider variety of input formats, including tar and zip archive formats.
A Filter Collection floe has been added that allows for filtering of existing collection.
The Giga Docking floe has a new docking mode Fast-FRED. Fast-FRED uses a simpler scoring function and samples slightly less in the initial stages of docking while using the same Chemgauss4 scoring function for final optimization and scoring. This docking mode can be x2-x4 times faster (and hence less costly) than the standard FRED. Results appear similar to standard FRED, but this docking mode has not been thoroughly validate as of this release in terms of quality of results.
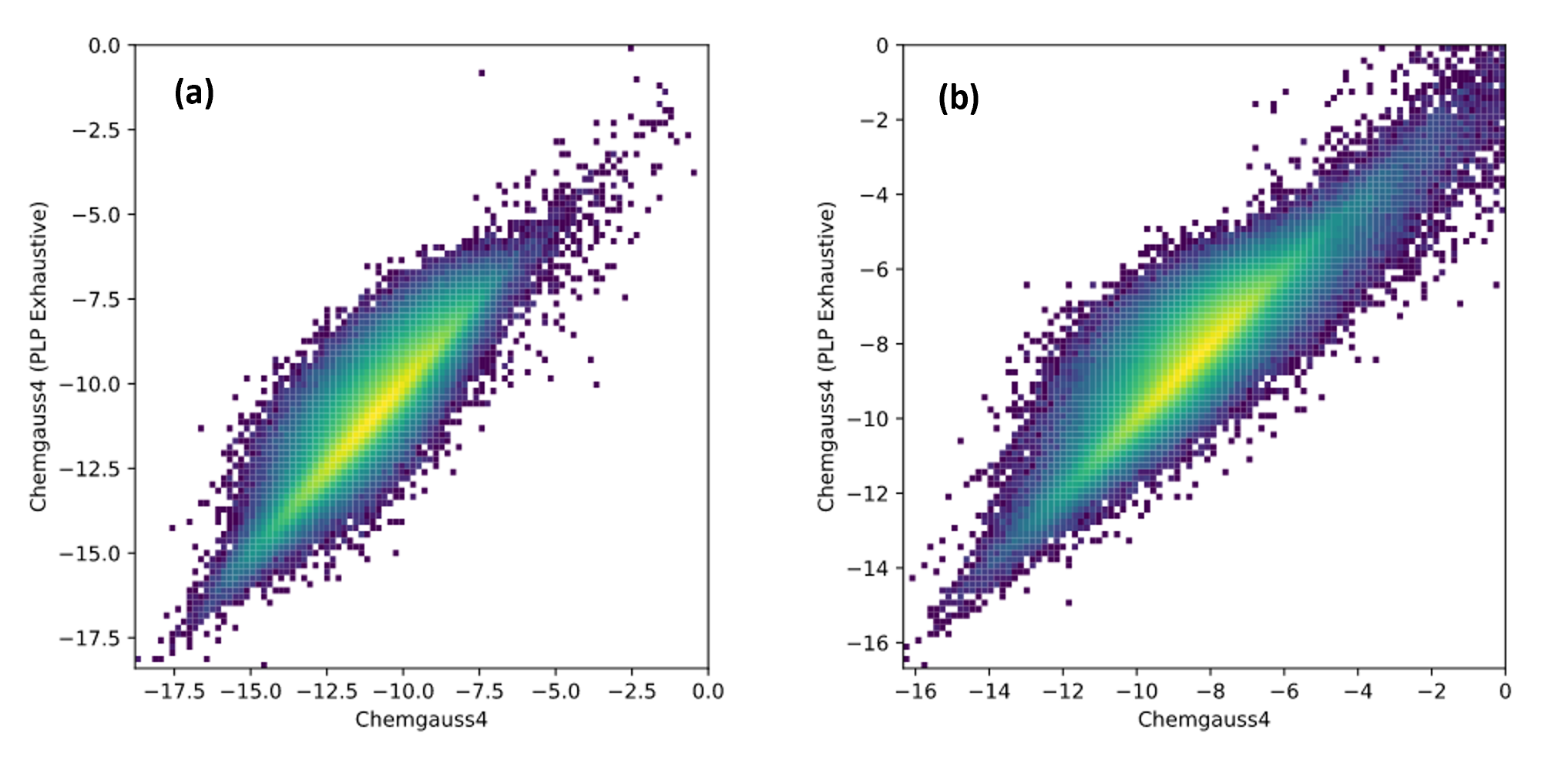
Comparison of FRED (x-axis) and Fast-FRED (y-axis) docking scores of 1M random Enamine Real molecules docked to (a) HSP90 and (b) PNPH
Fixes
An issue with the collection preparation floes duplicating some input molecules has been addressed.