OEToolkits 2015.Oct
FastROCS TK
OpenEye is pleased to announce that FastROCS is joining the pantheon of OpenEye toolkits as FastROCS TK. Shape technology continues to be at the center of OpenEye’s scientific mission, and the unprecedented performance gains of FastROCS TK continues to enable new science to be explored throughout the molecular modeling community.
Note
FastROCS TK is only available in Python on 64-bit Linux platforms.
|
|
|
Warning
If your FastROCS license was created before May 19th, 2015, please visit https://www.eyesopen.com/contact for a new license. FastROCS TK requires that the relevant “python” feature be turned on for access to FastROCS TK.
OEChem: Molecule Reading Performance Improvement
In addition to extending the capability of OpenEye’s toolkits, improving the performance of their existing functionalities is also important to us. We continually work to increase the performance of the molecule readers in OEChem TK to accommodate our field’s ever-growing molecule databases. Significant effort has been made to accelerate reading molecules into OEMol. The graph below illustrates the overall results of the code optimizations performed since the previous release (2015.Jun).
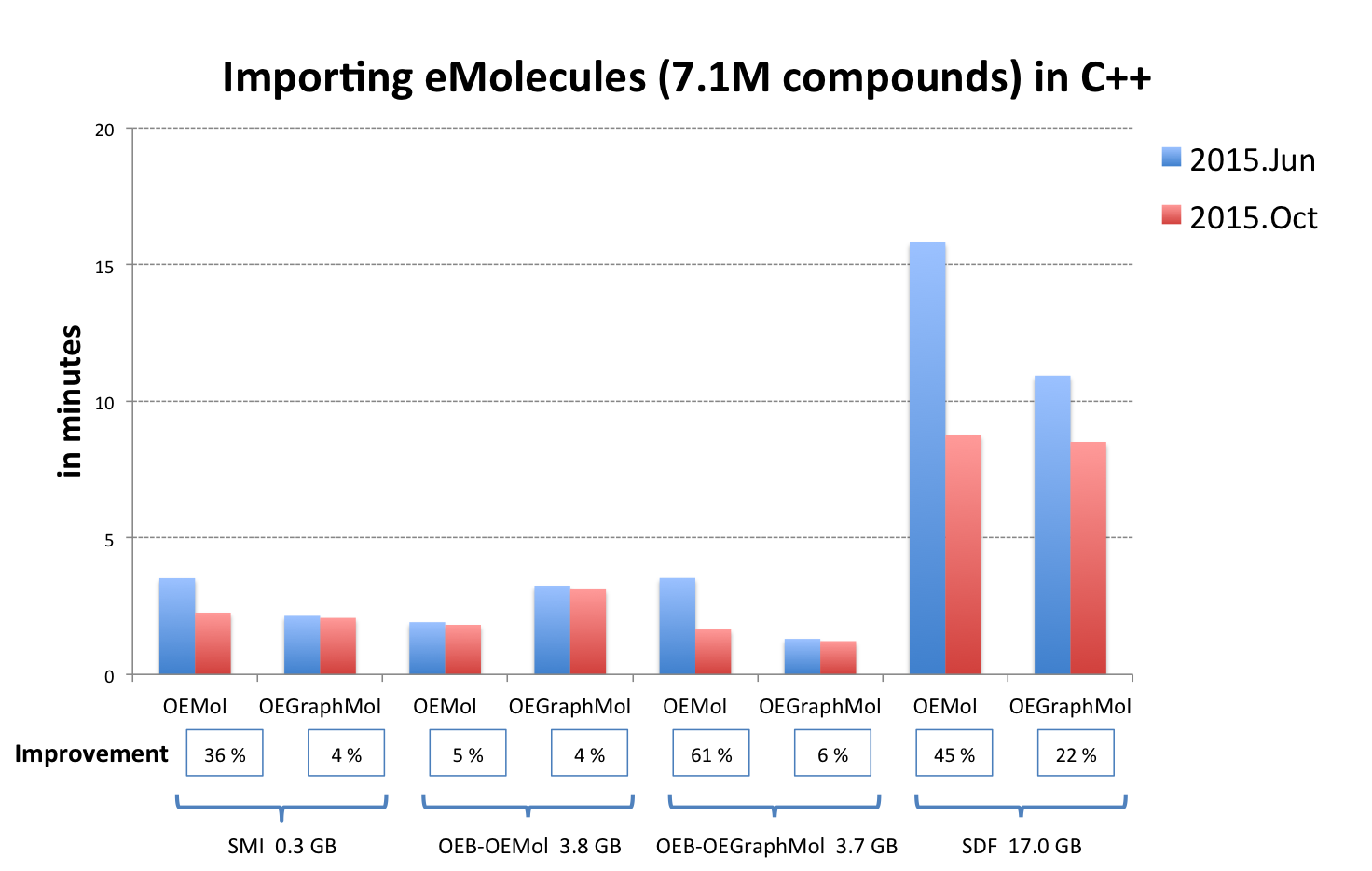
|
Note
OEB-OEMol - OpenEye OEBinary file format stores molecules as OEMol (multi-conformer molecule representation)
OEB-OEGraphMol - OpenEye OEBinary file format stores molecules as OEGraphMol (single-conformer molecule representation)
OEBio: Fragment Network
The 2015.Feb release introduced the visualization of receptor-ligand interactions by Grapheme TK. The following 2015.Jun release provided automatic identification of ligands by OEBio TK. This release extends these capabilities by providing read-only access to the fragment network data structure that can store interactions perceived by OEDocking TK. Several classes and predicates that provide convenient ways to examine various interactions types between a ligand and a protein have been also introduced.
For more details about using these new APIs, see the example in the Perceive and Print Protein-Ligand Interactions section.
General Notices
OpenEye Python Toolkits’ installation command has been changed to:
$ pip install -i https://pypi.anaconda.org/OpenEye/simple OpenEye-toolkits ... $ oecheminfo.py Installed OEChem version: |oechemversion| platform: ubuntu-14.04-g++4.8-x64 built: |builddate| ...
This is the last release to support FastROCS TK on Ubuntu 12.
The 2016.Feb release will be the last release to support Ubuntu 12 for all toolkits.
The 2015.Oct release is the last to support OSX 10.8.