OEToolkits 2018.Feb
GraphSim TK: GPU-Accelerated Fast Fingerprint Similarity Searches
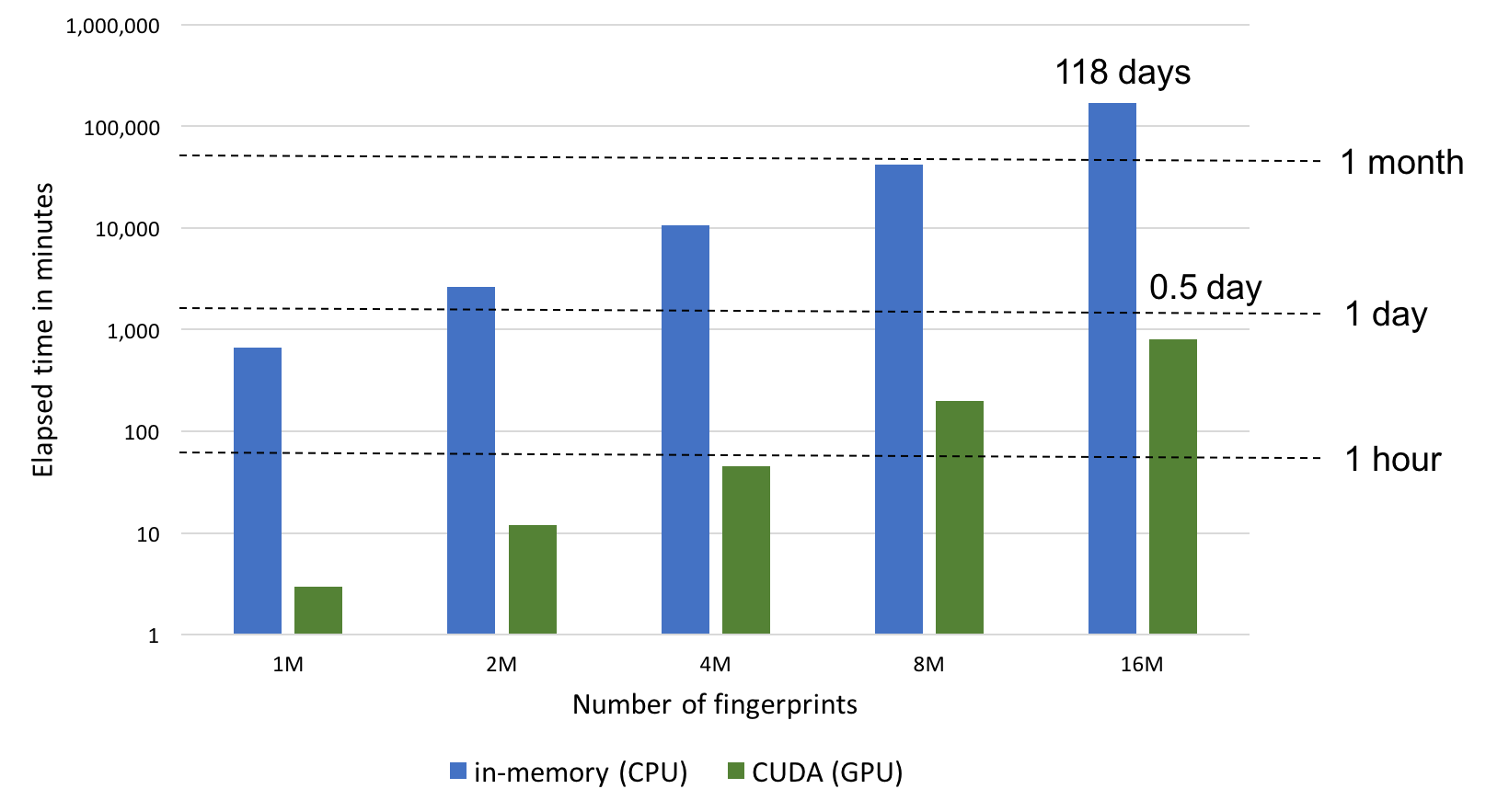
GraphSim TK now supports CUDA for even faster fingerprint searching, allowing users to perform molecular similarity calculations up to 200x faster than the in-memory fast fingerprint mode (see the NxN similarity score calculations graph below).
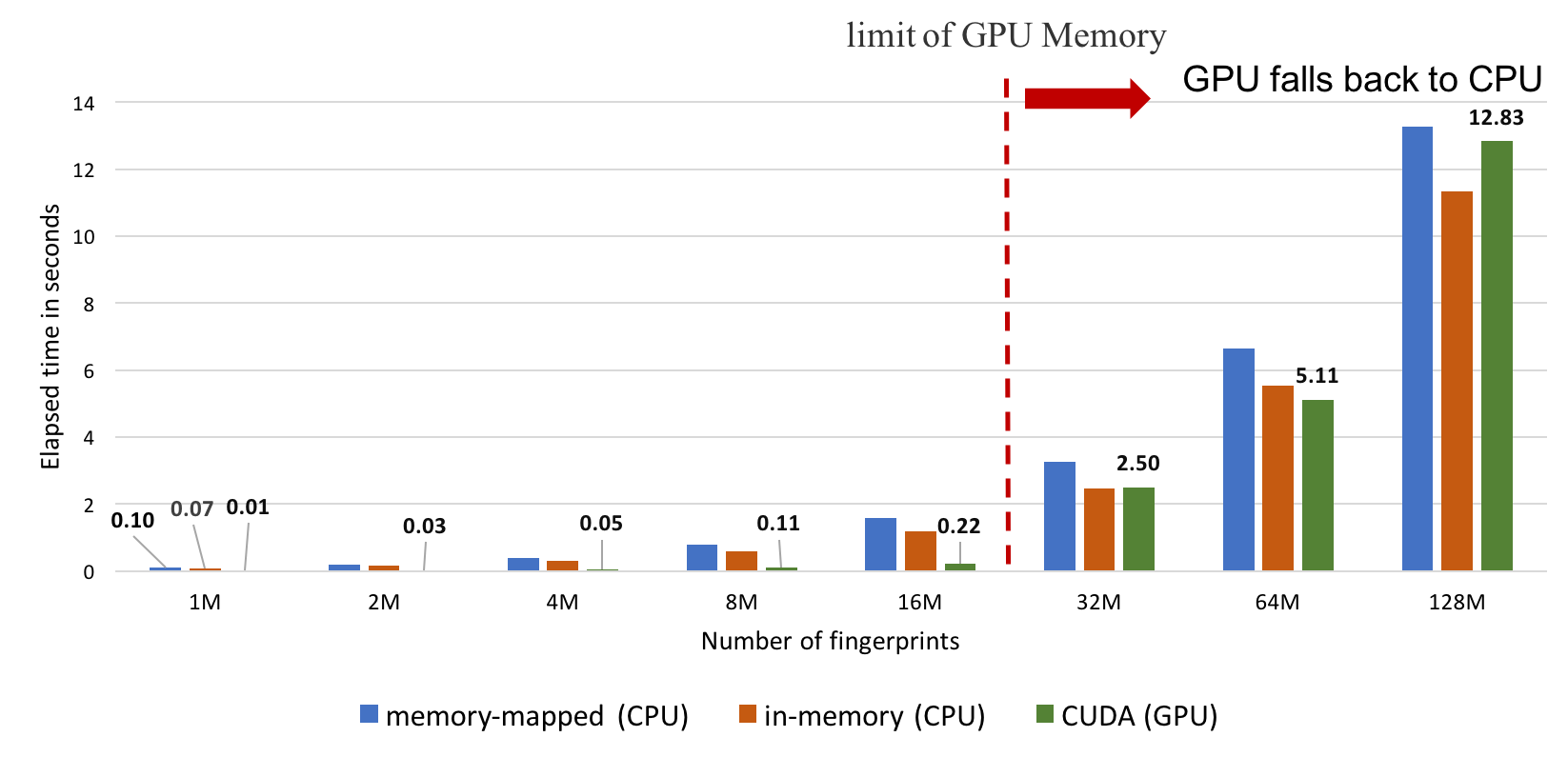
CUDA mode involves preloading all fingerprints into GPU memory prior to performing similarity calculations. Searches are limited by GPU memory availability and will fall back to the memory-mapped CPU mode if the entire set of fingerprints cannot be preloaded into the GPU memory. Despite this limitation, this represents the fastest way to perform similarity searches.
The most effective way to use CUDA fingerprint searches is to perform multiple searches with a pre-loaded database on a dedicated GPU machine. If CUDA mode is selected but a GPU is not present on the system, the search will simply fall back to memory-mapped mode.
The new OEFastFPDatabase::GetHistogram method
performs an NxN search of a database and returns a histogram of scores.
This is valuable information for understanding the diversity of a
database without consuming excess disk space.
Using OEFastFPDatabase::GetHistogram in CUDA
mode will afford 2.5 billion searches per second
(benchmarked on an AWS P3 instance - NVIDIA Tesla V100) compared with
12.5 million searches per second when using memory-mapped mode on the CPU.
See the GraphSim TK Examples chapter of the OEGraphSim TK documentation for more information and examples using the new OEFastFPDatabase class.
Grapheme TK : Unpaired Interaction Map
Grapheme TK now includes the ability to generate unpaired interaction maps (see the example below). An unpaired interaction map provides a complementary view to the more common active site interaction depiction. While the interaction map (on the right) depicts interactions between the ligand and protein, the unpaired map (on the left) illustrates interactions that could contribute to binding but are not formed in the complex. Together, these two maps of the protein-ligand binding site provide insights into protein-ligand interactions and communicate complex 3D structural results in a directly actionable way.
In the unpaired interaction map, different linker types are used to mark unpaired acceptor and donor hydrogen bond interactions (see the figure on the left). Note that since these interactions are unpaired, the direction of the linkers has no real spatial meaning. Ligand linkers are directed away from the ligand, while protein linkers are directed towards the ligand.
The geometric parameters used to perceive unpaired interactions are customizable to fit user needs.
Conda Packages for Python
The 2018.Feb release is the first release to fully support Conda packages
for Python toolkits on all supported platforms. The toolkits are available
as a package named openeye-toolkits in the openeye Conda channel on
the default conda.anaconda.org server. Versions are available for
Python 3.5 and higher on Windows and Python 3.4 and higher on Linux and macOS.
On Linux and macOS, users can install the toolkits into a Conda Python
environment with conda install -c openeye openeye-toolkits. On Windows,
Anaconda Navigator is a convenient way to manage Conda packages.
See Python Installation for more information.
General Notices
All OpenEye Toolkit Python code examples now follow the PEP8 standard.
The 2018.Feb release fully supports macOS High Sierra 10.13.
mac OS 10.10 is no longer supported.