Multi Page Images
Multi-page images can be generated by the usage of the
OEMultiPageImageFile class.
After generating a multi-page object by specifying the orientation and
the size of its pages, the individual pages can be accessed by calling
the OEMultiPageImageFile.NewPage method.
The returned OEImage then can be used to depict
molecule(s) as demonstrated in the previous chapters.
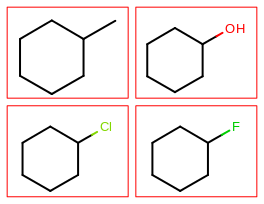
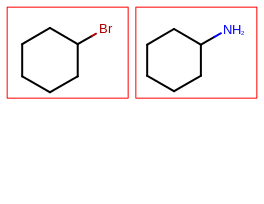
The image created by Listing 1 is shown in
Figure: Example multi-page depiction.
Listing 1: Example of multi-page depiction
public class MultiPage {
private static OEImageGrid MakeNewPageGrid(OEMultiPageImageFile multi) {
int rows = 2;
int cols = 2;
OEImage image = multi.NewPage();
OEImageGrid grid = new OEImageGrid(image, rows, cols);
grid.SetCellGap(20);
grid.SetMargins(20);
return grid;
}
public static void main(String argv[]) {
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("C1CC(C)CCC1");
smiles.add("C1CC(O)CCC1");
smiles.add("C1CC(Cl)CCC1");
smiles.add("C1CC(F)CCC1");
smiles.add("C1CC(Br)CCC1");
smiles.add("C1CC(N)CCC1");
OEMultiPageImageFile multi = new OEMultiPageImageFile(OEPageOrientation.Landscape,
OEPageSize.US_Letter);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions();
OEImageGrid grid = MakeNewPageGrid(multi);
OEImageBaseIter celliter = grid.GetCells();
for (String smi : smiles) {
if (!celliter.IsValid()) {
// go to new page
grid = MakeNewPageGrid(multi);
celliter = grid.GetCells();
}
OEImageBase cell = celliter.Target();
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smi);
oedepict.OEPrepareDepiction(mol);
opts.SetDimensions(cell.GetWidth(), cell.GetHeight(), OEScale.AutoScale);
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(cell, disp);
oedepict.OEDrawBorder(cell, oedepict.getOERedPen());
celliter.Increment();
}
oedepict.OEWriteMultiPageImage("MultiPage.pdf", multi);
}
}
page 1 |
page 2 |
|---|---|
|
|
The Listing 1 example creates a PDF multi-page image
file, the multi-page image types supported by OEDepict TK can be accessed by
calling the OEGetSupportedMultiPageImageFileExtensions
function.
It returns an iterator over the file extensions that can be used when
writing a multi-page image file by the
OEWriteMultiPageImage function.
Multi Page Reports
The Listing 1 example illustrates how depict a set of
molecules in a multi-page PDF using the OEImageGrid
and OEMultiPageImageFile classes.
OEDepict TK also provides the OEReport class to do
this task in a more convenient way.
The OEReport class is a multi-page layout manager that
handles the page generation and the positioning of information
(such as text and molecule depiction).
In the Listing 2 example, first a
OEReportOptions object is created that stores
all properties that determine the layout of a documentation.
After generating the OEReport object a
molecule can be positioned on a cell that is returned by
the OEReport.NewCell method.
The OEReport.NewCell method creates cells
from left to right and top to bottom order on each page.
If no more cells are left on the page, then a new page is created
before returning the first cell of this new page.
After the document is generated, i.e all molecule are depicted,
the OEWriteReport function writes the multi-page
documentation into into a file with the given name.
The image created by the Listing 2 example is the
same as depicted in Figure: Example multi-page depiction.
Listing 2: Example of multi-page depiction using the OEReport class
public class MultiPageReport {
public static void main(String argv[]) {
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("C1CC(C)CCC1");
smiles.add("C1CC(O)CCC1");
smiles.add("C1CC(Cl)CCC1");
smiles.add("C1CC(F)CCC1");
smiles.add("C1CC(Br)CCC1");
smiles.add("C1CC(N)CCC1");
int rows = 2;
int cols = 2;
OEReportOptions reportopts = new OEReportOptions(rows, cols);
reportopts.SetPageOrientation(OEPageOrientation.Landscape);
reportopts.SetCellGap(20);
reportopts.SetPageMargins(20);
OEReport report = new OEReport(reportopts);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(report.GetCellWidth(), report.GetCellHeight(), OEScale.AutoScale);
for (String smi : smiles) {
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smi);
oedepict.OEPrepareDepiction(mol);
OEImageBase cell = report.NewCell();
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(cell, disp);
oedepict.OEDrawBorder(cell, oedepict.getOERedPen());
}
oedepict.OEWriteReport("MultiPageReport.pdf", report);
}
}
See also
OEReportOptions class
OEReport class