Molecule Layouts
The previous chapters described how to depict a single molecule in an image. This chapter provides examples for drawing multiple molecular diagrams into the same image.
Depicting Molecules in an Arbitrary Position
The OEImageFrame class provides a general framework to display a molecule in a specific region of the image by defining the following parameters:
the reference of the parent image
the width and height of the generated frame
the offset of the frame relative to the top-left corner of its parent (OE2DPoint)
The Listing 1 examples generates an image where the same
molecule is depicted in different regions of the image.
The molecular diagrams are automatically resized in order to fit into
the specific frame, since the corresponding
OE2DMolDisplay object is constructed with the width
and height of the OEImageFrame object.
When the OERenderMolecule function is called, the
offset of the OEImageFrame object is added to the
coordinates stored in OE2DMolDisplay in order to
transfer the molecule into the specific region of the image.
The image created by Listing 1 is shown in
Figure: Example of using OEImageFrame.
Listing 1: Example of using OEImageFrame
public class ImageFrame {
private static void DrawMolecule(OEImageBase image, OEMolBase mol,
double width, double height, OE2DPoint offset) {
OEImageFrame frame = new OEImageFrame(image, width, height, offset);
oedepict.OEDrawBorder(frame, oedepict.getOELightGreyPen());
double scale = OEScale.AutoScale;
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(width, height, scale);
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
boolean clearBackground = true;
oedepict.OERenderMolecule(frame, disp, !clearBackground);
}
public static void main(String argv[]) {
OEImage image = new OEImage(400, 400);
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, "C1CC(O)CNC1");
oedepict.OEPrepareDepiction(mol);
DrawMolecule(image, mol, 60, 60, new OE2DPoint(50.0, 40.0));
DrawMolecule(image, mol, 180, 180, new OE2DPoint(180.0, 120.0));
DrawMolecule(image, mol, 80, 80, new OE2DPoint(300.0, 20.0));
DrawMolecule(image, mol, 50, 50, new OE2DPoint(150.0, 320.0));
DrawMolecule(image, mol, 20, 20, new OE2DPoint(360.0, 360.0));
oedepict.OEWriteImage("ImageFrame.png", image);
}
}
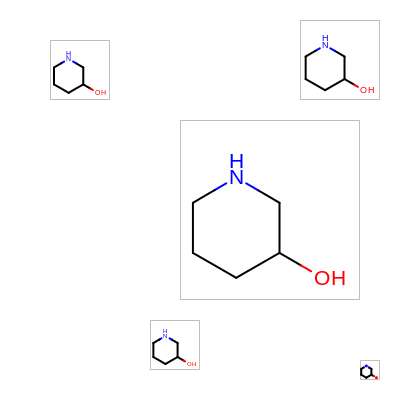
Example of using OEImageFrame
See also
OEImageFrame class
OEDrawBorderfunctionOERenderMoleculefunctionOEWriteImagefunction
Depicting Molecules in a Grid
The OEImageGrid class allows molecules to be aligned into
rows and columns.
After generating an OEImageGrid object by specifying its number
of rows and columns, the cells of the grid can be accessed (from left to right, top
to bottom order) by calling the OEImageGrid.GetCells method.
The image created by Listing 2 is shown in
Figure: Example of using OEImageGrid.
Listing 2: Example of using OEImageGrid
public class ImageGridSimple {
public static void main(String args[]) {
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("C1CC(C)CCC1");
smiles.add("C1CC(O)CCC1");
smiles.add("C1CC(Cl)CCC1");
OEImage image = new OEImage(200, 200);
int rows = 2;
int cols = 2;
OEImageGrid grid = new OEImageGrid(image, rows, cols);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(grid.GetCellWidth(),
grid.GetCellHeight(),
OEScale.AutoScale);
OEImageBaseIter celliter = grid.GetCells();
for (String smi : smiles) {
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smi);
oedepict.OEPrepareDepiction(mol);
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(celliter.Target(), disp);
celliter.Increment();
}
oedepict.OEWriteImage("ImageGridSimple.svg", image);
}
}
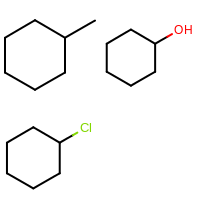
Example of using OEImageGrid
See also
OEImageGrid class
When an OE2DMolDisplay object is initialized, the dimensions of the depicted molecule is controlled by the width, height and scale parameters of the OE2DMolDisplayOptions object.
When only one molecule is depicted then auto scaling of this molecule is preferred
(i.e. scaling the molecule in order to maximally fit it to the given dimensions).
However, when more that one molecule, that are quite different in
sizes, are depicted next to each other, auto scaling of each molecule can be
visually misleading because it emphasizes molecules with greater scaling factors.
See example in Figure: Example of using OEImageGrid
that is generated by Listing 3.
Listing 3: Example of auto scaling of molecules
public class ImageGridAutoScale {
public static void main(String[] args) {
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("c1ccccc1");
smiles.add("c1cccc2c1ccnc2");
smiles.add("c1ccc2c(c1)cc3ccccc3n2");
OEImage image = new OEImage(400, 200);
int rows = 1;
int cols = 3;
OEImageGrid grid = new OEImageGrid(image, rows, cols);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(grid.GetCellWidth(),
grid.GetCellHeight(),
OEScale.AutoScale);
int counter = 0;
for (OEImageBase cell : grid.GetCells()) {
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smiles.get(counter));
oedepict.OEPrepareDepiction(mol);
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(cell, disp);
counter += 1;
}
oedepict.OEWriteImage("ImageGridAutoScale.svg", image);
}
}
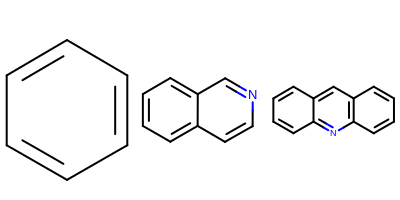
Example of auto scaling of molecules
See also
The following example (Listing 4) shows
how to ensure that all molecules are depicted using the same scaling
factor.
After initializing the molecules, the minimum scaling factor is
determined by looping over the molecules being depicted and calling
the OEGetMoleculeScale function.
This function returns the scaling factor of the molecule considering
the depiction options stored in the given
OE2DMolDisplayOptions object.
After determining the minimum scaling factor, the molecules are
rendered to the cells of the grid by using this fixed scaling factor.
The image created by Listing 4 is shown in
Figure: Example of using OEImageGrid.
Listing 4: Example of identical scaling of molecules
public class ImageGridFixedScale {
public static void main(String[] args) {
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("c1ccccc1");
smiles.add("c1cccc2c1ccnc2");
smiles.add("c1ccc2c(c1)cc3ccccc3n2");
ArrayList<OEGraphMol> mollist = new ArrayList<OEGraphMol>();
for (String smi : smiles) {
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smi);
oedepict.OEPrepareDepiction(mol);
mollist.add(mol);
}
OEImage image = new OEImage(400, 200);
int rows = 1;
int cols = 3;
OEImageGrid grid = new OEImageGrid(image, rows, cols);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(grid.GetCellWidth(),
grid.GetCellHeight(),
OEScale.AutoScale);
double minscale = Double.MAX_VALUE;
for (OEGraphMol mol : mollist) {
minscale = Math.min(minscale, oedepict.OEGetMoleculeScale(mol, opts));
}
opts.SetScale(minscale);
OEImageBaseIter celliter = grid.GetCells();
for (OEGraphMol mol : mollist) {
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(celliter.Target(), disp);
celliter.Increment();
}
oedepict.OEWriteImage("ImageGridFixedScale.svg", image);
}
}
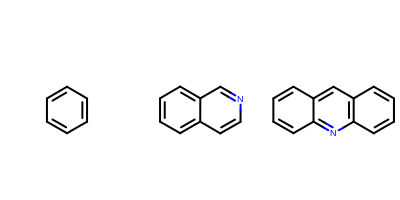
Example of identical scaling of molecules
See also
OEImageGrid.GetCellHeightmethodOEImageGrid.GetCellWidthmethod
Depicting Molecules in a Table
The following example demonstrates how to depict molecules and display data alongside them by using the OEImageTable. The individual cells of an OEImageTable object can be accessed by calling the any of the following methods:
These methods return an image (OEImageBase) on which
molecule can be drawn or text can be displayed using the
OEImageTable.DrawText method.
The image created by Listing 5 is shown in
Figure: Example of depicting molecules and data using image table.
Listing 5: Example of depicting molecules and data using image table
ArrayList<String> smiles = new ArrayList<String>();
smiles.add("c1ccccc1 benzene");
smiles.add("c1cccnc1 pyridine");
smiles.add("c1cncnc1 pyrimidine");
int imagewidth = 350;
int imageheight = 250;
OEImage image = new OEImage(imagewidth, imageheight);
int mainrows = 3;
int maincols = 2;
OEImageTableOptions maintableopts = new OEImageTableOptions(mainrows, maincols, OEImageTableStyle.NoStyle);
maintableopts.SetHeader(false);
ArrayList<Integer> colwidths = new ArrayList<Integer>();
colwidths.add(10);
colwidths.add(20);
maintableopts.SetColumnWidths(colwidths);
OEImageTable maintable = new OEImageTable(image, maintableopts);
int datarows = 4;
int datacols = 2;
OEImageTableOptions datatableopts = new OEImageTableOptions(datarows, datacols, OEImageTableStyle.LightGreen);
datatableopts.SetStubColumn(true);
datatableopts.SetMargins(5.0);
datatableopts.SetColumnWidths(colwidths);
for (int r = 0; r < mainrows; ++r) {
// depict molecule in first column
OEImageBase cell = maintable.GetBodyCell(r + 1, 1);
OE2DMolDisplayOptions opts = new OE2DMolDisplayOptions(cell.GetWidth(), cell.GetHeight(), OEScale.AutoScale);
opts.SetTitleLocation(OETitleLocation.Hidden);
OEGraphMol mol = new OEGraphMol();
oechem.OESmilesToMol(mol, smiles.get(r));
oedepict.OEPrepareDepiction(mol);
OE2DMolDisplay disp = new OE2DMolDisplay(mol, opts);
oedepict.OERenderMolecule(cell, disp);
// depicting data in table
cell = maintable.GetBodyCell(r + 1, 2);
OEImageTable datatable = new OEImageTable(cell, datatableopts);
datatable.DrawText(datatable.GetHeaderCell(1, false), "Property");
datatable.DrawText(datatable.GetHeaderCell(1), "Value");
datatable.DrawText(datatable.GetStubColumnCell(1), "Name");
datatable.DrawText(datatable.GetBodyCell(1, 1), mol.GetTitle());
datatable.DrawText(datatable.GetStubColumnCell(2), "SMILES");
datatable.DrawText(datatable.GetBodyCell(2, 1), oechem.OEMolToSmiles(mol));
datatable.DrawText(datatable.GetStubColumnCell(3), "MV");
String mwdata = String.format("%.3f", oechem.OECalculateMolecularWeight(mol));
datatable.DrawText(datatable.GetBodyCell(3, 1), mwdata);
}
oedepict.OEWriteImage("ImageTable.png", image);
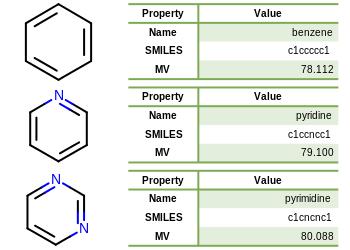
Example of depicting molecules and data using image table