Tutorial 2: How to prepare a database for faster load speeds
In this tutorial, you will learn how to pre-process a conformer database
file for FastROCS TK, allowing for faster database load times with
OEShapeDatabase::Open. Load times could be up to 10x
faster. See the figure below for an eMolecules dataset of 14 million
conformers.
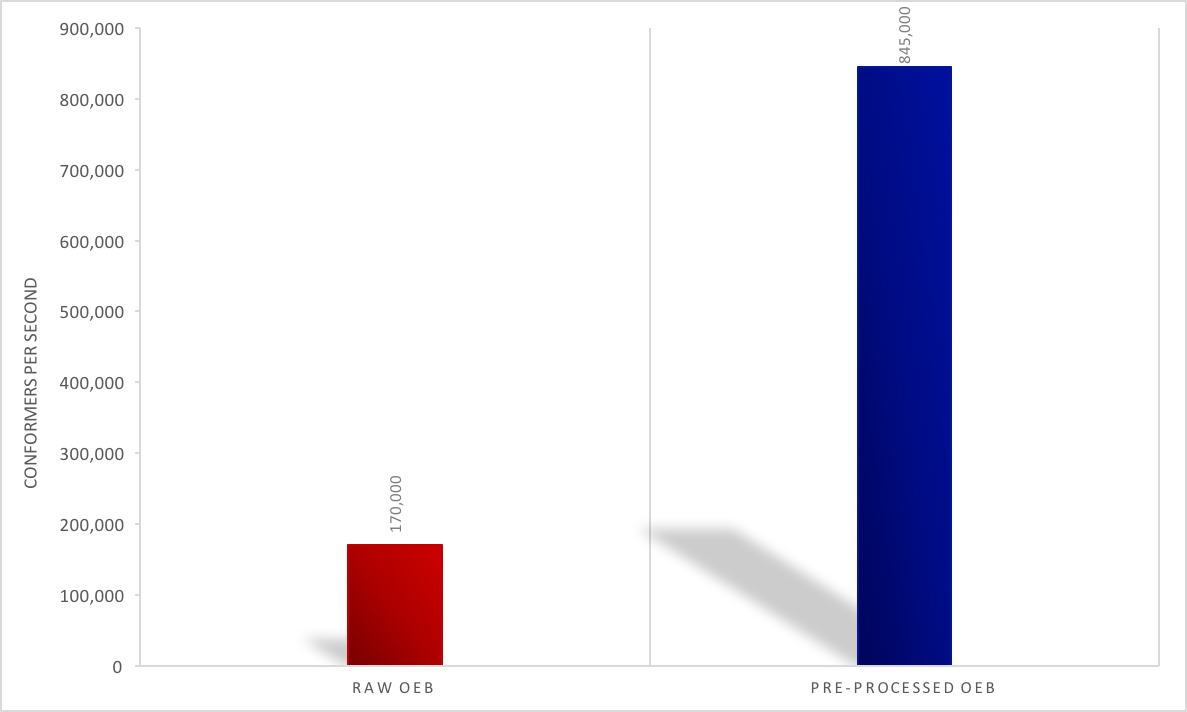
Pre-Processing Performance Impact
To gain this extra loading performance, you need to use the following functions:
OEPreserveRotCompress- this function works on the input molecule stream to ensure that rotor-offset compression is preserved during the preparation process. Rotor offset compression is a way of storing conformers as a set of torsions instead of storing the coordinates for every single conformer of a molecule. This optimization reduces the memory footprint of a multi-conformer molecule.OEPRECompress– this function works on the output molecule stream object allowing the molecules to be stored in a ‘pre-compressed’ format:Writes rotor-offset-compressed molecules in the perfect-rotor-encoding format
There is no need to Gzip which means faster
OEMolDatabase::Open.
OEPrepareFastROCSMol– this function works on each OEMol record of the input.oeb:Sets the energy of each conformer to 0.0 to avoid writing it to OEB.
Suppresses hydrogens and reorders reference conformers for compression.
Pre-calculates color atoms.
Pre-calculates self-color and self-shape terms for all conformers.
Note
The color terms cached by
OEPrepareFastROCSMolare from theOEColorFFType::ImplicitMillsDeancolor force field. A different color force field can be given as the second argument to override ImplicitMillsDean.
In general, calling OEPrepareFastROCSMol and
OEPRECompress will result in a smaller OEB file
than the default OEB.GZ output from OMEGA.
Further reduction in file-size can be achieved by using an
OEMCMolType::HalfFloatCartesian molecule to store
reference coordinates and torsions as 16-bit floating point.
Here is some example code showing how to pre-process a database with
OEPrepareFastROCSMol, save to a precompressed
format, and reduce the file size by using half precision:
For added convenience, we have created a shapedatabaseprep.cpp example script which can be modified to meet your exact needs:
Download code