Orion CLI
Options
The Orion CLI has several top level options that modify the behavior of the commands that are run.
Timeout
The --timeout flag can be used to set the request timeout in seconds, i.e.
if the server takes longer to respond to a request than the specified number of
seconds, the client disconnects. The default value is 10, but that may be too
short for some cases:
Slower or inconsistent internet connections
Sharing a package with many cubes to many users
Uploading large files
JSON
The --json flag can be used to have the Orion CLI return JSON
rather than plaintext to make it easier parse the results
ocli --json <command>
Profile
The --profile flag specifies which profile configuration to use while
making requests to the Orion API. The default is the default profile
ocli --profile=testing <command>
ocli config directory and file permissions are now forced to be read/write for the owner
Max Retries
The --max-retries flag specifies the number of times that the client
will retry a connection if it fails or gets a 404 error. This is set to 2
by default. There is back off implemented with the retries, so the time will
increase exponentially (to a certain point) with the number of retries.
Shard uploads and downloads are unaffected by --max-retries.
ocli --max-retries=5 <command>
Configuration Commands (ocli config)
Configure an Orion Profile
ocli config profile --url <hostname> --username <username> [--password <text>] [--with-token] [--project <project_id>]
Configure an Orion Profile for Use with SAML
To configure an Orion profile for use with SAML requires first creating a token in the web interface’s user profile page.
ocli config profile --url <hostname> --username <username> --with-token [--project <project_id>]
Change the Default Project of an Orion Profile
ocli config project
Delete an Orion Profile
ocli config delete <profile_name>
Retrieve Information about the Current Profile
ocli config info
List the Configured Profiles
ocli config list
Refresh Your Orion Profile token
ocli config token
Storage Commands
Folders
Commands to interact with Orion’s virtual filesystem
Note that folder commands are TOP LEVEL - e.g. ocli ls <path> instead of ocli folders ls <path>.
Folders contain Files, Datasets, ShardCollections, and other folders. They can take the following forms:
/project/<project-id>/user/<username>/<path>Project folders, which are private to user with the given username.
/project/<project-id>/team/<path>Project shared folders. All project members have access to these folders and their contents.
/project/<project-id>/workspace/<workspace-id>/<path>Project shared workspace folders. Project members are given permission by sharing the workspace with them.
/organization/<path>Organization shared folders.
List Contents of a Folder, or Details of an Item
Setting --recurse=True
will list all subdirectories and contents of the provided path(s). Items can be specified by path or
file://<file_id>|dataset://<dataset_id>.
ocli ls <path(s)|item(s)> [--recurse=bool]
Create a Folder
ocli mkdir <path(s)>
Copy to/from Orion
This works similarly to scp, but dest must be a folder or local directory.
Currently, only file and folder upload is supported. Setting --recurse=True will copy children
recursively. Setting --dry-run=True will list the items that would be copied and then quit.
ocli cp <source> <dest> [--recurse=<bool>] [--dry-run=<bool>]
Peek Inside a File or Dataset in Orion
Items can be specified by
path or by file://<file_id>|dataset://<dataset_id>. --number= or -n will optionally specify
the number of lines for a file or the number of records for a dataset.
ocli head <path(s)|item(s)> [-n, --number=<integer>]
Move a Folder or an Item
This will move all the children if the path refers to a Folder. Setting
--dry-run=True will list the move(s) and then quit.
Note: items will inherit the destination folder’s permissions. Therefore, mv can be used to share
across teams, workspaces, or organization.
ocli mv <source> <dest> [--dry-run=<bool>]
Remove a Folder or Item
Setting --dry-run=True will list the item(s) to be removed and then quit.
Note: currently will not remove a folder with children. In the future this will use the trash-can api.
ocli rm <path(s)> [--dry-run=<bool>]
Workspaces
The CLI provides a number of commands to interact with workspaces, shared areas for Orion files, datasets, and collections organized into folders.
Create a Workspace
To create a workspace, use the create command and give it a slug, a name unique to this project. The backend will automatically give the workspace a unique id, which can be used in subsequent calls to add or remove users by sharing the workspace.
ocli workspaces create <workspace_slug>
Datasets
The CLI provides several ways to interact with datasets
Get Information about a Dataset
Using the id of a dataset it is possible to access all of the information associated with the dataset
ocli datasets info <resource_id>
Delete a Dataset
To delete a dataset, use the following command
ocli datasets delete <resource_id>
List Datasets
To list the datasets the current Orion profile has access to. The arguments
passed after list are used as query parameters. For example
tags=2D,3D name=foo or name=aspirin.
ocli datasets list [--project=<integer>] [--name=<string>] \
[--limit=<integer>] \
[--offset=<integer>] \
[owner=<integer>] \
[deleted=<boolean>] \
[expired=<boolean>] \
[tags=TAG1,...] \
[tag_ids=<integer>,...] \
[systags=<tag>,...] \
[exclude_tags=<tag>,...] \
[tag_match=[default: exact](exact | iexact | contains | icontains | startswith | istartswith | endswith | iendswith)] \
[order_by=[-](created | name | count | owner)] \
[<key>=<value>,...]
Example:
ocli datasets list
ocli datasets list --name=aspirin_ --limit=30
ocli datasets list \
--project=12 \
--limit=20 \
tag_match=exact \
order_by=-size \
tags=3D,foo_jobs \
exclude_tags=2D,bar_jobs \
Upload a Dataset
To upload a dataset, pass the local path to the file and the client will create a new dataset. Handles any OEChem readable format, except JSON.
ocli datasets upload <file> [--project=<identifier>]
Download a Dataset
The following command will download a dataset, with any OEChem writable format except JSON, to your local file system.
ocli datasets download <resource_id> <file>
Finalize a Dataset
If the creation of a dataset is interrupted (for example, a job fails or is canceled as
it is writing records), then it will be left in a dirty state. The following
command will finalize a dataset, which sets dirty to false and provides
an accurate count of records contained in the dataset.
ocli datasets finalize <resource_id>
Files
The Orion CLI provides several ways to interact with Files.
Retrieve Information about an Orion File
ocli files info <resource_id>
Delete an Orion File
ocli files delete <resource_id>
List Orion Files
Possible to filter using query parameters.
For example tags=foo will filter Orion Files that have the tag foo.
ocli files list [--project=<integer>] [--name=<string>] \
[--limit=<integer>] \
[--offset=<integer>] \
[owner=<integer>] \
[deleted=<boolean>] \
[expired=<boolean>] \
[tags=TAG1,...] \
[tag_ids=<integer>,...] \
[tag_match=(exact | iexact | contains | icontains | startswith | istartswith | endswith | iendswith)] \
[order_by=[-](created | name | size | owner | id)] \
[<key>=<value>,...]
Examples:
ocli files list
ocli files list --name=foo_ --limit=10
ocli files list \
--project=12 \
--limit=20 \
tag_match=exact \
order_by=-size \
tags=mol_jobs,foo_jobs \
exclude_tags=bar_jobs \
Uploading a File from the Local File System to Orion
ocli files upload <file> [--project=<identifier>]
Downloading an Orion File to the Local File System
ocli files download <resource_id> <file>
Secrets
Secrets are a way of storing tokens in Orion. All values are encrypted upon upload and are decrypted when request through the API
Get Information on a Specific Secret
ocli secrets info <resource_id>
Delete an Existing Secret
ocli secrets delete <resource_id>
List the Secrets to Which You Have Access
ocli secrets list [--name=<string>] \
[--limit=<integer>] [--offset=<integer>] \
[description=<description>] [owner=<integer>] \
[<key>=<value>,...]
ex:
ocli secrets list
ocli secrets list --name=secret_foo
Create a New Secret
If --value is not provided, a prompt will provide the ability to set
the value using a password input to avoid storing the value in the terminal
history.
ocli secrets create <secret_name> [--value=<secret_value>] [--description=<string>]
Get Information on Sharing Status of a Specific Secret
ocli secrets list-shares <resource_id>
Projects
Commands for managing projects.
Get Information on a Specific Project
ocli projects info <resource_id>
Delete an Existing Project
ocli projects delete <resource_id>
List the Projects to Which You Have Access
ocli projects list [--limit=<integer>] [--offset=<integer>] \
[name=<name>] [as_admin=<boolean>] \
[deleted=<boolean>] [expired=<boolean>] \
[<key>=<value>,...]
ex:
ocli projects list
ocli projects list name=foo_
ocli projects list deleted=true --limit=5
Create a New Project
ocli projects create <project_name>
Leave a Project
ocli projects leave <project_id> [--new-owner=<user_id>]
Any member of a project can use this command to leave a project.
If
--new-ownerisn’t specified, the project assets belonging to the leaving user will be transferred to the project owner.If the user specified as
--new-ownerisn’t already a member of the project, they will automatically join the project.If the leaving user is the project owner
--new-ownermust be specified.
Get Information on Sharing Status of a Specific Project
ocli projects list-shares <resource_id>
Floe Commands
The following commands are a set of resources in Orion that relate to Floe which includes Packages, WorkFloes, and Jobs.
Packages
Commands related to Floe Packages
Initializing a Package
ocli packages init
The ocli packages init command:
Sets up a skeleton of an Orion package containing cubes and floes.
Provides simple example cube and floe.
Supports testing setup using PyTest including working tests for the example cube and floe.
Provides a
setup.pyfile with commands to run tests or build the package.Provides version configuration via storing the version only in the module’s
__init__.pyfile.
ocli packages
Upload and list Floe packages
Usage
ocli packages [OPTIONS] COMMAND [ARGS]...
check-image
Check an image for compatibility with Orion.
Usage
ocli packages check-image [OPTIONS]
Options
- -v, --verbose <verbose>
Set True to enable verbose output
create-environment-yaml
Convert a manifest.json file to an environment.yml.
Usage
ocli packages create-environment-yaml [OPTIONS] MANIFEST_PATH
Options
- -o, --output-path <output_path>
Specifies an output file to write to
Arguments
- MANIFEST_PATH
Required argument
delete
Delete a specific package.
Usage
ocli packages delete [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
deprecate
Deprecate a package and its contents.
Usage
ocli packages deprecate [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
download
Download Floe package source to the local file system.
Usage
ocli packages download [OPTIONS] IDENTIFIER FILE
Arguments
- IDENTIFIER
Required argument
- FILE
Required argument
environment
View the environment build log for a package
Usage
ocli packages environment [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
export-base-image
Export a base image to an Orion File
Usage
ocli packages export-base-image [OPTIONS] BASE_IMAGE
Options
- -o, --output-path <output_path>
Orion Folder to put exported image file in
- --notify, --no-notify
Set True to receive an email notification when the export is complete
- --download, --no-download
Wait for export to complete and download the result
Arguments
- BASE_IMAGE
Required argument
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli packages help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
import-from-orion-file
Create a Floe package from an Orion file
Usage
ocli packages import-from-orion-file [OPTIONS] IMAGE_FILE
Options
- -d, --delete-image-file <delete_image_file>
Specifies whether the package image file should remain stored in Orion after inspection
Arguments
- IMAGE_FILE
Required argument
import-from-registry
Create a Floe package by pulling an image from a registry
Ex: ocli packages import-from-registry –registry-url “my-team-registry/package-xyz-images” –image-tag package-xyz-v1 –pull-username mydockerhubuser –secret 83293
Usage
ocli packages import-from-registry [OPTIONS]
Options
- -r, --registry-url <registry_url>
Required URL of image registry, Docker Hub only requires <my_registry>/<my_repository>
- -t, --image-tag <image_tag>
Required Tag to pull from registry, ex: latest, package-xyz-v1
- -u, --pull-username <pull_username>
Required Image registry username
- -s, --secret <secret>
Required ID of Orion secret containing the password for the image registry
info
Retrieve information about a specific package
Usage
ocli packages info [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
init
Create a Floe package scaffold.
This command can be used to create the structure necessary for a Floe package, but is distinct from the set of package commands which can be used to manage package images.
Usage
ocli packages init [OPTIONS]
Options
- --full-name <full_name>
Required Author of the Floe package
- --email <email>
Required Email of the Floe package author
- --project-name <project_name>
Required Name of the project
- --project-slug <project_slug>
Required Short name for the project
- --module-name <module_name>
Required Name of the module
- --description <description>
Required Description of the project
- --version <version>
Required Package version
- --service
Include a service directory and boilerplate code
inspection
View the inspection log for a package.
Usage
ocli packages inspection [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
list
List Floe packages
Usage
ocli packages list [OPTIONS] [FILTERS]...
Options
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
Arguments
- FILTERS
Optional argument(s)
list-base-images
List base images available
Usage
ocli packages list-base-images [OPTIONS]
show-dockerfile
Shows an example Dockerfile to be used for a package image.
Usage
ocli packages show-dockerfile [OPTIONS]
Options
- -o, --output-path <output_path>
Specifies an output file to write to
show-packaging-help
Shows a short guide to building a package image.
Usage
ocli packages show-packaging-help [OPTIONS]
tag
Tag a specific package.
Usage
ocli packages tag [OPTIONS] IDENTIFIER [TAGS]...
Options
- --project <project>
Arguments
- IDENTIFIER
Required argument
- TAGS
Optional argument(s)
undeprecate
Un-deprecate a package and its contents.
Usage
ocli packages undeprecate [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
untag
Untag a specific package.
Usage
ocli packages untag [OPTIONS] IDENTIFIER [TAGS]...
Arguments
- IDENTIFIER
Required argument
- TAGS
Optional argument(s)
upload
Upload a Floe package from the local filesystem or URL.
Files are limited to 2GB in size. If a Floe package is larger, then please upload using a URL and ensure that your manifest.json has appropriate disk_space set in build_requirements. See the Manifest section of Floe’s Orion Integration documentation.
Usage
ocli packages upload [OPTIONS] PACKAGE_PATH
Arguments
- PACKAGE_PATH
Required argument
upload-package-image
Create a Floe package from an image file
Usage
ocli packages upload-package-image [OPTIONS] IMAGE_FILE
Options
- -p, --path <path>
Specifies an Orion folder for storing the uploaded image file
- -d, --delete-image-file <delete_image_file>
Specifies whether the package image file should remain stored in Orion after inspection
Arguments
- IMAGE_FILE
Required argument
Retrieving the Information about a Floe Package
ocli packages info <resource_id>
Retrieving the Environment Build Log for a Floe Package
ocli packages environment <resource_id>
Retrieving the Inspection Information for a Floe Package
ocli packages inspection <resource_id>
Deleting a Floe Package
ocli packages delete <resource_id>
Listing Floe Packages
ocli packages list [--limit=<integer>] [--offset=<integer>] \
[states=<state>,... (ready, deprecated, queued, error)] \
[name=<name>] [UUID=<uuid>] [owner=<integer>] \
[<key>=<value>,...]
ex:
ocli packages list
ocli packages list states=error
ocli packages list --limit=20 states=ready,queued
Upload a Floe Package
ocli packages upload <file_or_url>
Get Information on Sharing Status of a Specific Floe Package
ocli packages list-shares <resource_id>
WorkFloes
Commands for accessing Workfloes in Orion. Workfloes are a collection of cubes that defines the flow of data.
Retrieving the information about a WorkFloe
ocli workfloes info <resource_id>
Deleting a Workfloe
ocli workfloes delete <resource_id>
Listing Workfloes
ocli workfloes list [--package=<integer>] [--name=<string>] \
[--limit=<integer>] [--offset=<integer>] \
[states=<state>,... (ready, deprecated, hidden, invalid)] \
[systags=<tag>,...] [tags=<tag>,...] [owner=<integer>] \
[classification=<string> [classification_depth=<integer>]
[order_by=(recently_used | name)]
[<key>=<value>,...]
ex:
ocli workfloes list
ocli workfloes list tags=tag1,tag2
ocli workfloes list --name=collection_to_ --limit=10 order_by=recently_used
ocli workfloes list --package=10 states=ready,deprecated
Listing Classifications
ocli workfloes list-classifications [--name=<string>] [--path=<string>] \
[--depth=<integer>] [--limit=<integer>] [--offset=<integer>] \
[<key>=<value>,...]
ex:
ocli workfloes list-classifications
ocli workfloes list-classifications --path=Example/Test
ocli workfloes list-classifications --name=exam --depth=1 --limit=10
Get Information on Sharing Status of a Specific WorkFloe
ocli workfloes list-shares <resource_id>
Jobs
Commands for interacting with Jobs in Orion. Jobs are an instantiation of a Workfloe that performs the computation defined by the Workfloe.
Retrieving the Information about a Job
ocli jobs info <resource_id>
Retrieve the Logs of a Job
ocli jobs logs <resource_id>
Canceling a Running Job
ocli jobs cancel <resource_id>
Deleting a Job
ocli jobs delete <resource_id>
Displaying Size of Logs
Retrieve the size of the logs of an Orion job.
ocli jobs log_size [OPTIONS] JOB_ID
Arguments
- JOB_ID
Required argument identifying the job.
Deleting Job Logs
Delete the logs of an Orion job.
ocli jobs delete_logs [OPTIONS] JOB_ID
Arguments
- JOB_ID
Required argument
Listing Existing Jobs
ocli jobs list [--limit=<integer>] [--offset=<integer>] \
[name=<name>] [project=<integer>] [workfloe=<integer>] \
[job_type=(system | user)] [owner=<integer>] \
[states=<state>,... (queued, running, complete)] \
[tags=<tag>,...] [<key>=<value>,...]
ex:
ocli jobs list
ocli jobs list workfloe=20
ocli jobs list tags=tag1,tag2 states=complete
ocli jobs list job_type=user name=aspirin_ states=queued,running
Tag
Tag a specific job.
ocli jobs tag [OPTIONS] IDENTIFIER [TAGS]...
Options
–project <project>
Arguments
- IDENTIFIER
Required argument identifying the job.
- TAGS
Optional string argument(s) to be used as tags.
The –project` option applies the tags using project ID.
For example:
ocli jobs tag --project 25412 203761 candidates
Tagged WorkFloeJob 203761 with ('candidates',)
Project tags are displayed in pink in the user interface. Job tags are yellow.
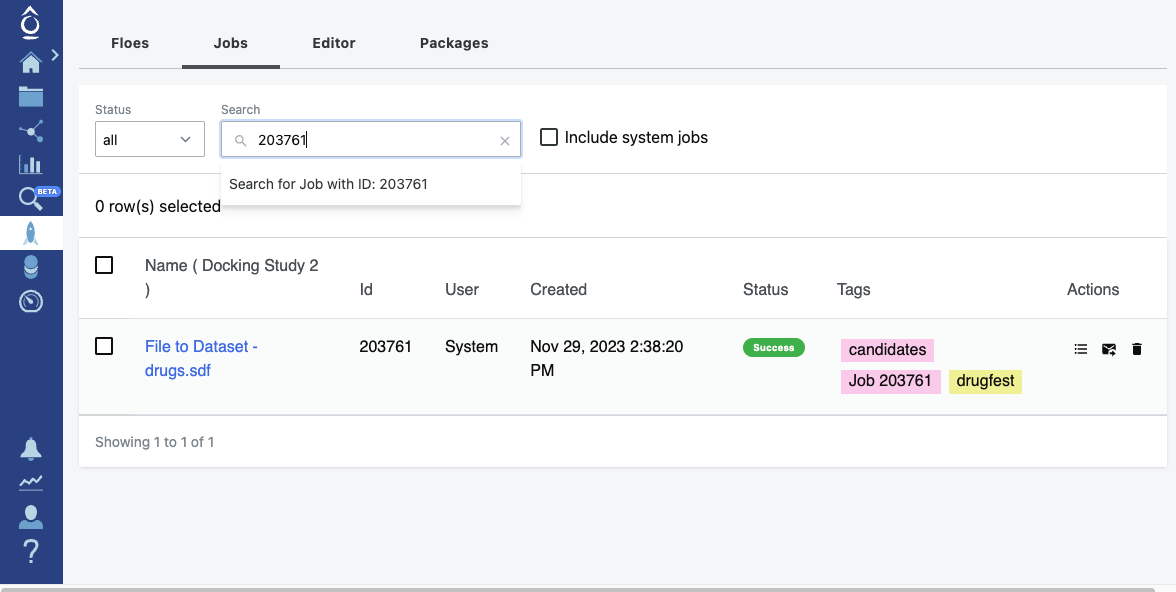
Project Tags (Pink) and Job Tags (Yellow)
Both kinds of tag can be used in searches in the user interface.
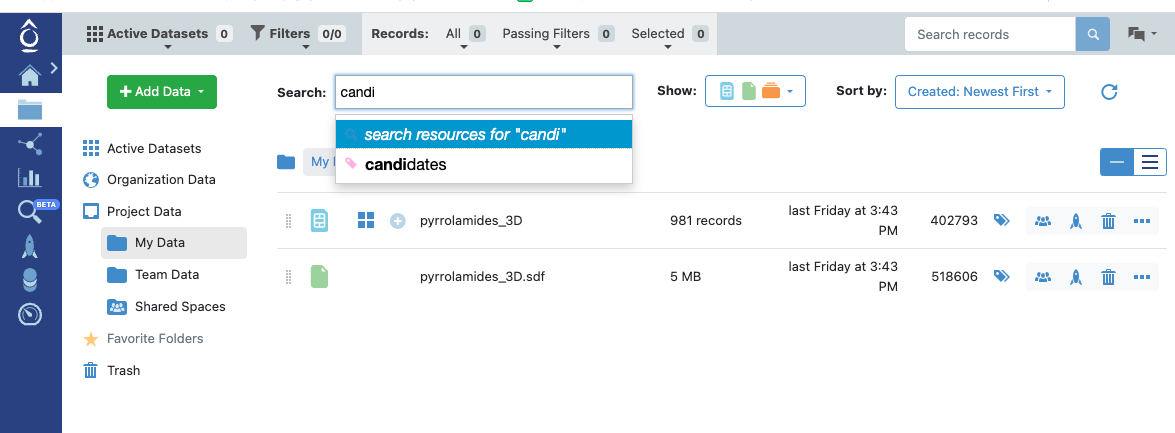
Searching by a Project Tag Value
Starting a Job
ocli jobs start <workfloe_id> <job_name> [--wait] \
[FLOE_PARAMETERS]
ex:
ocli jobs start 6836 job_name0 --in 601 --out run_output.oedb
Note
Adding -- --help to the ocli jobs start command will print the
parameters that are expected by the Workfloe.
Watching a Job
ocli jobs watch <workfloe_id> <job_name> [CUBES] [OPTIONS]
Collect Debug Information
ocli jobs debug-export <job_id>
Retrieving Summary of Jobs Within an Organization
ocli jobs summary-export [--package-uuids=<str>] [--workfloe-uuid=<str>] \
[--job_ids=<str>] [--start-date=<str>] [--end-date=<str>] \
[--output-path=<str>] [--download=(--download | --no-download)] \
[--notify=(--notify | --no-notify)]
Examples:
ocli jobs summary-export
ocli jobs summary-export --notify --download
ocli jobs summary-export --start-date 2023-01-01 --end-date "2023-01-01 18:00:00"
ocli jobs summary-export --output-path "/project/<project-id>/users/<user-id>/folder-name/"
ocli jobs summary-export --package-uuids "uuid_A, uuid_B" --workfloe-uuids "uuid_C" --job-ids "6, 19, 47"
This functionality is available only to stack admins, org admins and mt-org admins.
Cost Exporting
Generating Storage Use Report
The report generated by the following commands contains estimates based on the amount of storage used. They do not reflect the exact billed cost.
The results do not take into account the AWS tiered storage system.
Generating the report does not move anything from a lower cost tier into a higher cost tier.
ocli cost export --report-type=storage_use [USER IDS]
ex:
ocli cost export --report-type=storage_use 10 11 20
Generating Spend Summary Report
This report provides per-user totals for storage, jobs, and services. Costs for user created Molecule Search databases are included in the services total. Totals are pulled from the ledger entries and aggregated over the previous 7 days (168 hours), current calendar month, and previous calendar month. If no user IDs are provided, data for the current user will be reported (non-admin) or for all organization users (admin).
ocli cost export --report-type=spend_summary [USER IDS]
ex:
ocli cost export --report-type=spend_summary 10 11 20
The resulting csv has column headers providing content labels, but the schema is not versioned, so for programmatic usage, the orionclient LedgerEntry is recommended. All amounts are in dollars.
ocli cost export
Triggers cost report in Orion, optionally downloads the file.
notify: Send an email upon completion.
output-path: The output path to store the cost report in.
report-type: The report type
redact-project-names: Redact project names from reports
users: A list of user ids for user data to include in the cost report.
Example:
ocli cost export –notify=true –report-type=spend_summary 10
ocli cost export –report-type=spend_summary 10 11 20
ocli cost export –report-type=storage_use 10 11 20
Usage
ocli cost export [OPTIONS] [USERS]...
Options
- --notify <notify>
Notify upon completion via email
- --output-path <output_path>
Project path
- --report-type <report_type>
Required Report type
- Options:
storage_use | spend_summary
- --redact-project-names <redact_project_names>
Redact project names
- --download, --no-download
Wait for export to complete and download the result
Arguments
- USERS
Optional argument(s)
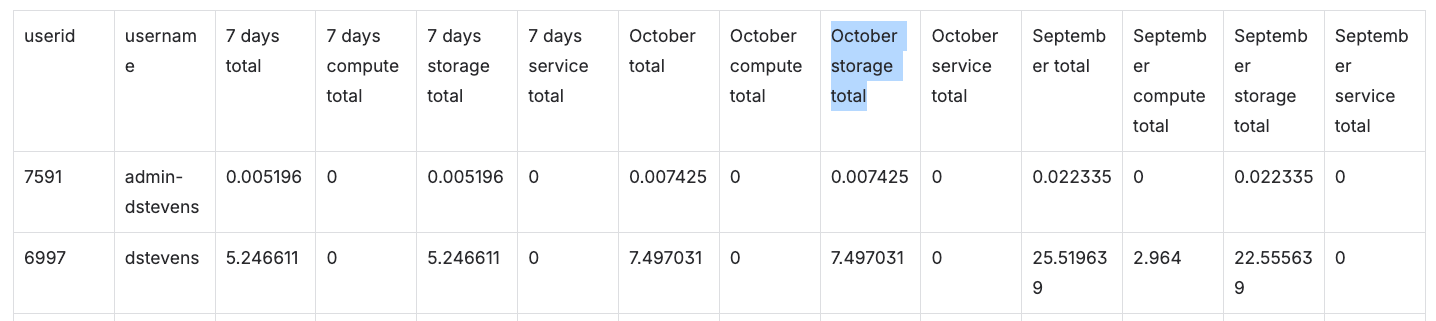
Sample Cost Export Table
Molecule Search Database Commands
These commands are used to manage molecule search databases.
ocli molsearch db
Orion molecule search database commands.
Usage
ocli molsearch db [OPTIONS] COMMAND [ARGS]...
create
Submit a job to create a molecule search database.
Usage
ocli molsearch db create [OPTIONS]
Options
- --collection <collection>
Collection with molecule search database.
- --cpu <cpu>
Number of CPUs.
- --memory-mb <memory_mb>
Memory (in MB).
- --disk-space <disk_space>
Disk (in GB).
- --gpu <gpu>
Number of GPUs.
- --instance-type <instance_type>
Instance type.
- --pools <pools>
Pools to run the job in.
- --search-type <search_type>
Search type ‘2D’ or ‘3D’
- Options:
2D | 3D
- --server-is-remote
Is the server being manually managed?
- --server-address <server_address>
Address of the remote server.
- --auth-secret <auth_secret>
Authorization secret of the remote server.
- --verify-ssl-cert
Should the remote server verify its SSL certificate?
- --display-name <display_name>
User provided display name for the database
- --section <section>
Value of database section - OE provided or Customer Managed
- --is-proxy
Flag to use proxy or not
delete
Delete a molecule search database.
Usage
ocli molsearch db delete [OPTIONS] DATABASE
Arguments
- DATABASE
Required argument
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli molsearch db help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
info
Information about a specific molecule search database.
Usage
ocli molsearch db info [OPTIONS] DATABASE
Arguments
- DATABASE
Required argument
list
List all molecule search databases.
Usage
ocli molsearch db list [OPTIONS] [FILTERS]...
Options
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
Arguments
- FILTERS
Optional argument(s)
load
Load a molecule search database.
Usage
ocli molsearch db load [OPTIONS] DATABASE
Arguments
- DATABASE
Required argument
logs
Retrieve the logs of a molecule search database
Usage
ocli molsearch db logs [OPTIONS] IDENTIFIER
Options
- --from <from_dt>
start date for the logs
- --until <until>
end date for the logs
Arguments
- IDENTIFIER
Required argument
unload
Unload a molecule search database.
Usage
ocli molsearch db unload [OPTIONS] DATABASE
Arguments
- DATABASE
Required argument
update
Update a molecule search database.
Usage
ocli molsearch db update [OPTIONS] DATABASE
Options
- --section <section>
Value of database section - OE provided or Customer Managed
- --display-name <display_name>
User provided display name for the database
Arguments
- DATABASE
Required argument
Molecule Search Query Commands
These commands are used to manage molecule search queries.
ocli molsearch query
Orion molecule search query commands.
Usage
ocli molsearch query [OPTIONS] COMMAND [ARGS]...
cancel
Cancel a substructure molecule search query.
Usage
ocli molsearch query cancel [OPTIONS] QUERY
Arguments
- QUERY
Required argument
create
Create a molecule search query.
Usage
ocli molsearch query create [OPTIONS] COMMAND [ARGS]...
exact
2D exact search using a SMILES string. If a file is provided, it will be converted into smiles.
Usage
ocli molsearch query create exact [OPTIONS] DATABASE QUERY
Options
- --search-type <search_type>
How to search.
- Options:
ISM | ABS | ISOMORPH | UNCOLOR
- --name <name>
- --max-hits <max_hits>
- --wait
- --project <project>
Arguments
- DATABASE
Required argument
- QUERY
Required argument
fastrocs
3D similarity search using a fastrocs query
Usage
ocli molsearch query create fastrocs [OPTIONS] DATABASE QUERY_MOL_FILE_PATH
Options
- --name <name>
- --max-hits <max_hits>
- --shape-only
- --sim-type <sim_type>
- Options:
tanimoto | tversky
- --tversky-alpha <tversky_alpha>
- --orientation <orientation>
- Options:
inertial | inertialAtHeavyAtoms | inertialAtColorAtoms | subrocs | random
- --random-starts <random_starts>
- --wait
- --project <project>
Arguments
- DATABASE
Required argument
- QUERY_MOL_FILE_PATH
Required argument
graphsim
2D similarity search using a GraphSim query. If a file is provided, it will be converted into smiles.
Usage
ocli molsearch query create graphsim [OPTIONS] DATABASE QUERY
Options
- --fpname <fpname>
Fingerprint to use.
- --cutoff <cutoff>
Cutoff.
- --measure <measure>
Similarity measure to use.
- Options:
Tanimoto | Tversky | Dice | Cosine
- --name <name>
- --max-hits <max_hits>
- --wait
- --project <project>
Arguments
- DATABASE
Required argument
- QUERY
Required argument
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli molsearch query create help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
substructure
2D substructure search using an OEChem substructure query.
Usage
ocli molsearch query create substructure [OPTIONS] DATABASE INPUT_QUERY
Options
- --subsearch_query_type <subsearch_query_type>
- Options:
MDL | SMARTS
- --aliphatic-constraint
only applicable for MDL queries
- --topology-constraint
only applicable for MDL queries
- --stereo-constraint
only applicable for MDL queries
- --isotope-constraint
only applicable for MDL queries
- --name <name>
- --max-hits <max_hits>
- --wait
- --cancel-after <cancel_after>
- --project <project>
Arguments
- DATABASE
Required argument
- INPUT_QUERY
Required argument
title
Search a 2D database using a space separated list of titles.
Usage
ocli molsearch query create title [OPTIONS] DATABASE [TITLE]...
Options
- --name <name>
- --project <project>
Arguments
- DATABASE
Required argument
- TITLE
Optional argument(s)
delete
Delete a molecule search query.
Usage
ocli molsearch query delete [OPTIONS] QUERY
Arguments
- QUERY
Required argument
download
Download molecule search results and query to separate local files.
Downloaded query file will be named <result_file_name>-query.<ext>
Usage
ocli molsearch query download [OPTIONS] QUERY FILE
Options
- --ids <ids>
Comma-separated list of result IDs to export.
- --exclude
Exclude ids from results.
Arguments
- QUERY
Required argument
- FILE
Required argument
export
Export search results to a dataset.
The optional --path will place the export output in
a non-default location, as with:
--path /project/<project-id>/user/<username>/<custom_path>/
if unset, this value defaults to: /project/<project-id>/user/<username>/
The optional --project will place the export output in
a non-default project, as with:
--project <project-id>
A list of currently-available projects can be found with:
ocli projects list
Usage
ocli molsearch query export [OPTIONS] QUERY
Options
- --path <path>
Orion path where exported dataset is placed
- --project <project>
- --name <name>
Name of output dataset.
- --format <format>
Output format.
- --background
Export in async mode?
- --ids <ids>
Comma-separated list of result IDs to export.
Arguments
- QUERY
Required argument
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli molsearch query help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
info
Information about a specific molecule search query.
Usage
ocli molsearch query info [OPTIONS] QUERY
Arguments
- QUERY
Required argument
list
List all molecule search queries.
Usage
ocli molsearch query list [OPTIONS]
Options
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
- --query-type <query_type>
Type of query to list
- --project <project>
- --programmatic <programmatic>
Filter queries created by ocli (True), UI (False), or all queries (None)
list-results
List results for a single molecule search query.
Usage
ocli molsearch query list-results [OPTIONS] QUERY
Options
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
Arguments
- QUERY
Required argument
smiles
Get SMILES string for the molecule with the given title.
Usage
ocli molsearch query smiles [OPTIONS] TITLE
Arguments
- TITLE
Required argument
update
Update a molecule search query.
Usage
ocli molsearch query update [OPTIONS] QUERY
Options
- --name <name>
Query name.
- --saved <saved>
Save query.
Arguments
- QUERY
Required argument
Services
Orion Services is a registry of internal or external services integrated with the larger Orion system.
Service Specifications
These commands create and manipulate service specifications.
ocli services spec
Specification of Orion Service.
Orion services is a registry of internal or external services integrated with the larger Orion system.
Usage
ocli services spec [OPTIONS] COMMAND [ARGS]...
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli services spec help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
info
Retrieve an Orion service specification
Usage
ocli services spec info [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
list
List available specifications of Orion services
Usage
ocli services spec list [OPTIONS] [FILTERS]...
Options
- --name <name>
Filter by name
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
Arguments
- FILTERS
Optional argument(s)
Service Tasks
These commands create and manipulate service tasks from service specifications.
ocli services task
Management of Orion Services.
Orion services is a registry of internal or external services integrated with the larger Orion system.
Usage
ocli services task [OPTIONS] COMMAND [ARGS]...
create
Create an Orion service
Usage
ocli services task create [OPTIONS]
Options
- --name <name>
- --description <description>
- --specification <specification>
- --external <external>
- --url <url>
delete
Delete an Orion service
Usage
ocli services task delete [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
help
Long format of help CMD, or the first level commands if CMD is not specified.
Usage
ocli services task help [OPTIONS] [CMD]...
Arguments
- CMD
Optional argument(s)
info
Retrieve an Orion service
Usage
ocli services task info [OPTIONS] IDENTIFIER
Arguments
- IDENTIFIER
Required argument
list
List Orion services
Usage
ocli services task list [OPTIONS] [FILTERS]...
Options
- --name <name>
Filter by name
- --offset <offset>
Offset from the start of the list
- --limit <limit>
Number of objects to list
Arguments
- FILTERS
Optional argument(s)
logs
Retrieve the logs of a service task
Usage
ocli services task logs [OPTIONS] IDENTIFIER
Options
- --from <from_dt>
start date for the logs
- --until <until>
end date for the logs
Arguments
- IDENTIFIER
Required argument
update
Update an Orion service
Usage
ocli services task update [OPTIONS] IDENTIFIER
Options
- --name <name>
- --description <description>
- --state <state>
Arguments
- IDENTIFIER
Required argument