Depicting CSV or SDF in XLSX (Excel)¶
Problem¶
You want to depict molecules along with their associated data read from a CSV file in an xlsx Excel file. See example in drugs.xlsx and in Figure 1.
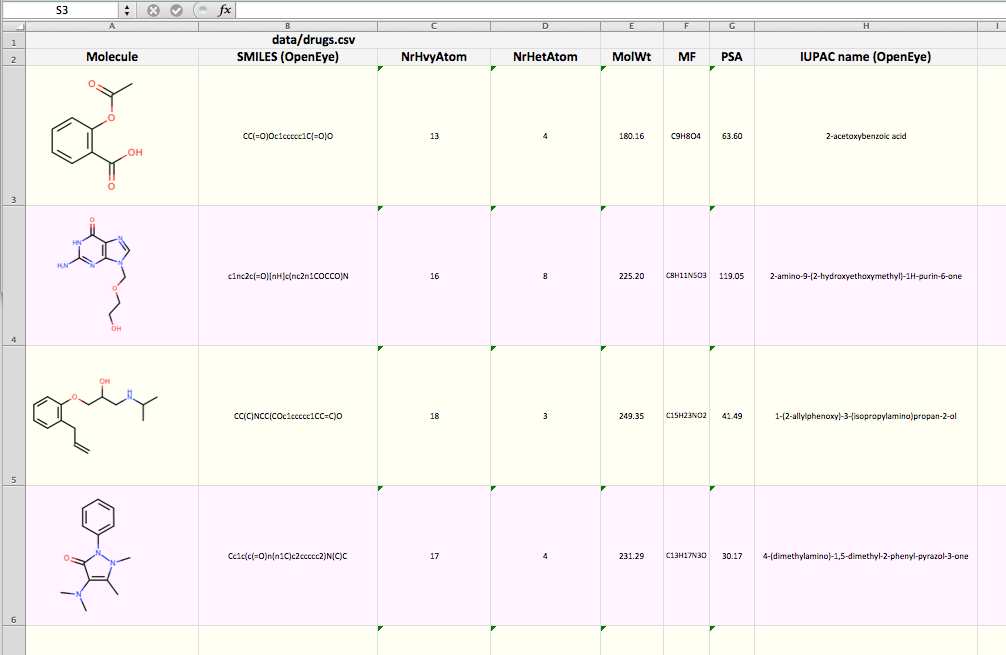
Figure 1. Example of depicting CSV in XLSX (The screen-shot is reduced here for visualization convenience)
Ingredients¶
|
Solution¶
The CSV file format is a text file format containing comma-separated values. In OEChem TK this file format is implemented to enable data exchange with a wide variety of other software. Each line of a CSV file stores data for a molecule that is represented by a SMILES string.
See also
- CSV File Format section of the OEChem TK documentation about the layout of the CSV file format.
When reading a CSV file, the fields of the file are attached to each molecule as SD data. This data can be accessed by the OEGetSDDataIter function that returns an iterator over all the SD data (tag - value) pairs of a molecule. The CollectDataTags function iterates over a list of molecules and returns the unique tags of the data attached to the molecules.
1 2 3 4 5 6 7 8 9 | def CollectDataTags(mollist):
tags = []
for mol in mollist:
for dp in oechem.OEGetSDDataIter(mol):
if not dp.GetTag() in tags:
tags.append(dp.GetTag())
return tags
|
The WriteXLSXFile function takes a list of molecules read from a CSV file along with the data tags returned by the CollectDataTags function.
First a ‘worksheet’ is created and styles are added by calling the AddHeadFormat and AddDataFormats functions (lines 5-9). Then the width of each column of the spreadsheet is estimated using the OEEstimateTextWidth function (lines 13-21). This is followed by generating the header of the the table by adding the input filename into the first row (lines 25-27) and each of the data tags into the second (lines 29-37). Then iterating over the molecules, each molecule is depicted in a new row along with the corresponding data with alternating styles. In order to add the images, a temporary image file has to be generated for each molecule. These files can be removed after the workbook is closed (lines 66-71).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 | def WriteXLSXFile(oname, mollist, iname, tags, opts):
# create an Excel file and add formating
workbook = xlsxwriter.Workbook(oname)
worksheet = workbook.add_worksheet()
headfont, headformat = AddHeadFormat(workbook)
datafont, dataformat_even, dataformat_odd = AddDataFormats(workbook)
# estimate width of columns
maxwidths = []
maxwidths.append(opts.GetWidth() * 0.15)
for tag in tags:
maxwidth = oedepict.OEEstimateTextWidth(tag, headfont) * 2.0
for mol in mollist:
if oechem.OEHasSDData(mol, tag):
value = oechem.OEGetSDData(mol, tag)
estimatedwidth = oedepict.OEEstimateTextWidth(value, datafont)
maxwidth = max(maxwidth, estimatedwidth)
maxwidths.append(maxwidth * 0.12)
# generate header
row, col = 0, 0
worksheet.set_row(row, None, headformat)
worksheet.merge_range('A1:D1', iname)
row, col = 1, 0
worksheet.set_row(row, None, headformat)
worksheet.set_column(col, col, maxwidths[col])
worksheet.write(row, col, "Molecule")
for tag in tags:
col += 1
worksheet.set_column(col, col, maxwidths[col])
worksheet.write(row, col, tag)
# generate rows
tmpfnames = []
for mol in mollist:
row += 1
if row % 2 == 0:
dataformat = dataformat_even
else:
dataformat = dataformat_odd
worksheet.set_row(row, opts.GetHeight() * 0.75, dataformat)
col = 0
fname = "tmp%d.png" % row
WriteImageToFile(fname, mol, opts)
worksheet.insert_image(row, col, fname)
tmpfnames.append(fname)
for tag in tags:
col += 1
value = "N/A"
if oechem.OEHasSDData(mol, tag):
value = oechem.OEGetSDData(mol, tag)
worksheet.write(row, col, value)
workbook.close()
# remove temporary image files
for fname in tmpfnames:
os.remove(fname)
|
The WriteImageToFile function generates a molecule depiction and writes it into an image file.
1 2 3 4 5 | def WriteImageToFile(fname, mol, opts):
oedepict.OEPrepareDepiction(mol)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule(fname, disp, False)
|
The AddHeadFormat function adds a style to the ‘workbook’ that is used in the header of the table.
1 2 3 4 5 6 7 8 9 10 11 | def AddHeadFormat(workbook):
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Bold, 18,
oedepict.OEAlignment_Center, oechem.OEBlack)
format = workbook.add_format({'bold': True, 'align': 'center',
'valign': 'vcenter', 'size': 18})
format.set_bg_color('#F4F4F4')
format.set_border_color('#DDDDDD')
format.set_border()
return font, format
|
The AddDataFormats function adds two formats to the ‘workbook’ one for the even and one for the odd rows of the table.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 | def AddDataFormats(workbook):
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Default, 12,
oedepict.OEAlignment_Center, oechem.OEBlack)
format_even = workbook.add_format({'bold': False, 'align': 'center',
'valign': 'vcenter', 'size': 12})
format_even.set_shrink()
format_even.set_bg_color('#FFFFF4')
format_even.set_border_color('#DDDDDD')
format_even.set_border()
format_odd = workbook.add_format({'bold': False, 'align': 'center',
'valign': 'vcenter', 'size': 12})
format_odd.set_shrink()
format_odd.set_bg_color('#FFF4FF')
format_odd.set_border_color('#DDDDDD')
format_odd.set_border()
return font, format_even, format_odd
|
Download code
csv2xlsx.py and drugs.csv supporting data
Usage
Running the above command will generate the drugs.xlxs file.
prompt > python3 csv2xlsx.py drugs.csv drugs.xlsx
Discussion¶
Reading the columns of an CSV file into SD data fields, means that the OEChem TK provides a meta-data interchange between sdf files and CSV files. Consequently, the same Python script can be used to generate a xlsx file reading an sdf file.
Usage
After downloading drugs.sdf supporting data file, the above command will generate the same drugs.xlsx file (apart from the input filename on the top).
prompt > python3 csv2xlsx.py drugs.sdf drugs.xlsx
See also¶
See also in OEChem TK manual¶
Theory
- SD Tagged Data Manipulation section
- CSV File Format section
API
- OEGetSDDataPairs function
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEPrepareDepiction function
