Depicting Multiple Matches¶
Problem¶
You want to perform multiple substructure searches and highlight the matches on the hit molecules. See example in Table 1.
| page 1 | page 2 |
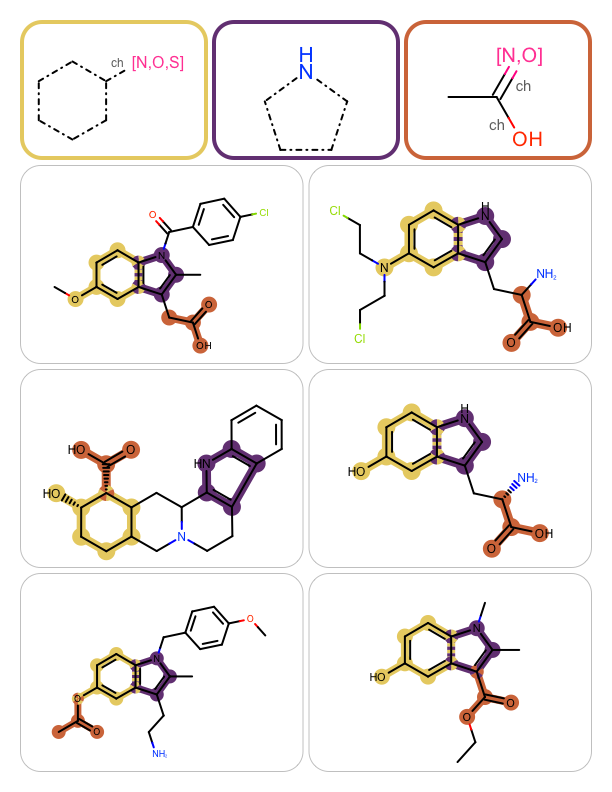
|
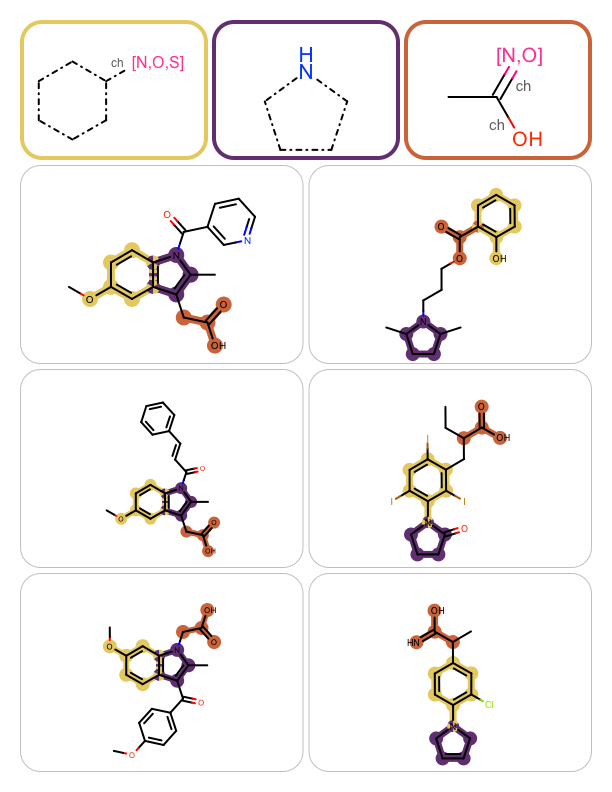
|
Ingredients¶
|
Solution¶
The GetSubstructureSearch function shows how to read MDL query files (OEReadMDLQueryFile) and initialize a OEQMol object by calling the OEBuildMDLQueryExpressions function. This OEQMol object is then used to initialize the OESubSearch object that performs substructure searches. Setting the maximum number of matches to 1 (see line 21) ensures the search will terminate upon finding one match. The GetSubstructureSearch function returns both the query molecule that will be depicted in the top of each page of the report and the substructure search object.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 | def GetSubstructureSearch(queryfname):
qifs = oechem.oemolistream()
if not qifs.open(queryfname):
oechem.OEThrow.Fatal("Cannot open mdl query file!")
if qifs.GetFormat() != oechem.OEFormat_MDL:
oechem.OEThrow.Fatal("Query file has to be an MDL file!")
querymol = oechem.OEGraphMol()
if not oechem.OEReadMDLQueryFile(qifs, querymol):
oechem.OEThrow.Fatal("Cannot read query molecule!")
oedepict.OEPrepareDepiction(querymol)
qmol = oechem.OEQMol()
queryopts = oechem.OEMDLQueryOpts_Default | oechem.OEMDLQueryOpts_SuppressExplicitH
oechem.OEBuildMDLQueryExpressions(qmol, querymol, queryopts)
subsearch = oechem.OESubSearch()
if not subsearch.Init(qmol):
oechem.OEThrow.Fatal("Cannot initialize substructure search!")
subsearch.SetMaxMatches(1)
return (querymol, subsearch)
|
The GetSubstructureSearches function iterates over a list of query file names and collects the query molecules and the substructure search objects (returned by the GetSubstructureSearch function) in two separate lists.
1 2 3 4 5 6 7 8 9 10 11 | def GetSubstructureSearches(queryfnames):
querymols = []
subsearches = []
for queryfname in queryfnames:
querymol, subsearch = GetSubstructureSearch(queryfname)
querymols.append(oechem.OEGraphMol(querymol))
subsearches.append(oechem.OESubSearch(subsearch))
return querymols, subsearches
|
The DepictMoleculesWithSubstructureMatches function that depicts the hits of the substructure searches takes the following parameters:
- report
- The OEReport object that allows the generation of multi-page documentation.
- mollist
- The list of target molecules.
- subsearches
- The list of OESubSearch objects initialized by the GetSubstructureSearches function.
- opts
- The OE2DMolDisplayOptions object that defines the style of the molecule depiction.
- colors
- The list of colors that is used to highlight the substructure search matches.
DepictMoleculesWithSubstructureMatches iterates over the target molecules and performs substructure searches by calling the GetSubstructureMatches function that:
- returns an empty list, if the target molecule does not contain all substructures or
- returns the list of substructure matches, one match for each successful substructure search
In the later case, the molecule (i.e. the hit) is rendered into the next cell of the report and the matched substructures are highlighted by calling the OEAddHighlightOverlay function. The OEAddHighlightOverlay takes all matches being highlighted and colors the overlapped atoms and bonds using the colors by turn. The colors used for highlighting are determined when the OEHighlightOverlayByBallAndStick object is constructed (see line 3).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | def DepictMoleculesWithSubstructureMatches(report, mollist, subsearches, opts, colors):
highlight = oedepict.OEHighlightOverlayByBallAndStick(colors)
for mol in mollist:
matches = GetSubstructureMatches(subsearches, mol)
if len(matches) == 0: # at least one substructure search fails
continue
oedepict.OEPrepareDepiction(mol)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OEAddHighlightOverlay(disp, highlight, matches)
cell = report.NewCell()
oedepict.OERenderMolecule(cell, disp)
oedepict.OEDrawCurvedBorder(cell, oedepict.OELightGreyPen, 20)
|
The GetSubstructureMatches function iterates over the substructure searches. If a substructure search fails (see lines 7-8) then the function returns an empty list. After each successful search the match list is appended with a new match stored in an OEAtomBondSet object.
1 2 3 4 5 6 7 8 9 10 11 12 13 | def GetSubstructureMatches(subsearches, mol):
unique = True
matches = []
for ss in subsearches:
miter = ss.Match(mol, unique)
if not miter.IsValid():
return []
else:
match = miter.Target()
matches.append(oechem.OEAtomBondSet(match.GetTargetAtoms(), match.GetTargetBonds()))
return matches
|
After generating the report with the substructure search matches the queries can be depicted on each page of the report. The DepictQueries function iterates over the headers of the OEReport object and depicts each query in a row that is generated using an OEImageGrid object. A border is drawn around each query molecule with its associated color to aid finding the corresponding substructure matches in the hit molecules.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | def DepictQueries(report, queries, colors):
for header in report.GetHeaders():
grid = oedepict.OEImageGrid(header, 1, len(queries))
grid.SetCellGap(4)
cellwidth, cellheight = grid.GetCellWidth(), grid.GetCellHeight()
opts = oedepict.OE2DMolDisplayOptions(cellwidth, cellheight, oedepict.OEScale_AutoScale)
colors.ToFirst()
for cell, query, color in zip(grid.GetCells(), queries, colors):
disp = oedepict.OE2DMolDisplay(query, opts)
oedepict.OERenderMolecule(cell, disp)
pen = oedepict.OEPen(oechem.OEWhite, color, oedepict.OEFill_Off, 4.0)
oedepict.OEDrawCurvedBorder(cell, pen, 20)
|
Download code
mdlsearches2pdf.py and supporting data files: query-A.mol, query-B.mol, query-C.mol, and targets.ism
Usage:
prompt > python3 mdlseacrhes2pdf.py -queries query-A.mol query-B.mol query-C.mol -target targets.ism -report matches.pdf
Discussion¶
Using colors with high contrast is recommended when highlighting overlapping matches by the OEAddHighlightOverlay function. In this example the colors returned by the OEGetContrastColors function are used.

Figure 1: Colors of maximum contrast returned by the OEGetContrastColors function
Even though there is no limit on the number of overlapping patterns that can be highlighted simultaneously by the OEAddHighlightOverlay function, attempting to highlight too many patterns will result in a complex image that will be difficult to visually interpret (see example in Figure 2).
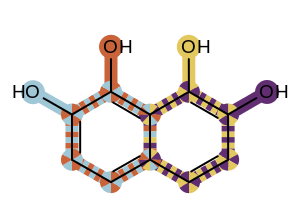
Figure 2: Example of highlighting extremely overlapping patterns
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
- Highlighting Overlapped Patterns section
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEAddHighlightOverlay function
- OEHighlightOverlayByBallAndStick class
- OEImage class
- OEImageGrid class
- OEPrepareDepiction function
- OERenderMolecule function
- OEReport class
