Depicting Fragment Contributions of XLogP¶
Problem¶
You want to depict the contribution of fragments to the total XLogP on your molecule diagram. See example in Figure 1.
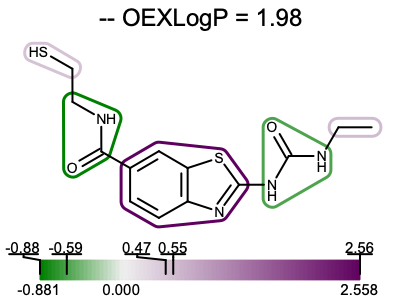
Figure 1. Example of depicting the fragment contributions of XLogP
Ingredients¶
|
Download¶
Download code
fragxlogp2img.py and fragxlogp2pdf.py
See also the Usage (fragxlogp2img) and Usage (fragxlogp2pdf) subsections.
Solution¶
The code snippet below shows how to calculate the total XLogP of a molecule along with the atom contributions by calling the OEGetXLogP function. Each atom contribution is then attached to the relevant atom as generic data with the given tag.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 | def set_atom_properties(mol, datatag):
"""
Attaches the XLogP atom contribution to each atom with the given tag.
:type mol: oechem.OEMolBase
:type datatag: string
"""
oequacpac.OERemoveFormalCharge(mol)
avals = oechem.OEFloatArray(mol.GetMaxAtomIdx())
logp = oemolprop.OEGetXLogP(mol, avals)
mol.SetTitle(mol.GetTitle() + " -- OEXLogP = %.2f" % logp)
for atom in mol.GetAtoms():
val = avals[atom.GetIdx()]
atom.SetData(datatag, val)
|
The fragment_molecule function fragments a molecule by using either the OEGetRingChainFragments, the OEGetRingLinkerSideChainFragments or the OEGetFuncGroupFragments function. Each enumerated fragment is then added to the molecule as a new group with the given tag.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | def fragment_molecule(mol, fragfunc, grouptag):
"""
Fragments the molecule and stores each fragment as a group on the molecule.
:type mol: oechem.OEMolBase
:type fragfunc: function()
:type grouptag: string
"""
for frag in fragfunc(mol):
atoms = oechem.OEAtomVector()
for atom in frag.GetAtoms():
atoms.append(atom)
bonds = oechem.OEBondVector()
for bond in frag.GetBonds():
bonds.append(bond)
mol.NewGroup(grouptag, atoms, bonds)
|
The set_fragment_properties function should be called after the atom contributions are calculated (see set_atom_properties) and the molecule is fragmented (see fragment_molecule). It iterates over the fragments, adds together the atom contributions and attaches this accumulated value to each group (fragment). It also returns the minimum and maximum fragment contribution.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 | def set_fragment_properties(mol, datatag, grouptag):
"""
Calculates the fragment contribution based on attached atom properties
for pre-generated fragments.
:type mol: oechem.OEMolBase
:type datatag: string
:type grouptag: string
"""
minvalue = float("inf")
maxvalue = float("-inf")
for group in mol.GetGroups(oechem.OEHasGroupType(grouptag)):
sumprop = 0.0
for atom in group.GetAtoms():
sumprop += atom.GetData(datatag)
group.SetData(datatag, sumprop)
minvalue = min(minvalue, sumprop)
maxvalue = max(maxvalue, sumprop)
return minvalue, maxvalue
|
The depict_molecule_fragment_xlogp function below shows how to project the fragment contributions of the total XLogP into a 2D molecular diagram. First the atom contributions are calculated by calling the set_atom_properties function (see lines 13-15). Then a molecule is fragmented (lines 19-21) and the fragment contributions are calculated (line 25). Using the minimum and maximum fragment contributions a color gradient is constructed (lines 29-32). Since both the molecule and the color gradient will be depicted, the image has to be divided into two image frames (lines 36-40). The molecule display is then initialized (lines 44-45) along with the highlighting style (lines 49-50) and the display option for the color gradient (line 52). Then fragment contributions are visualized by iterating over them and highlighting them on the molecule diagram using the color corresponding to their contributions (lines 54-64). Finally, the molecule along with the color gradient is rendered to the image (lines 68-69). You can see the result in Figure 1.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 | def depict_molecule_fragment_xlogp(image, mol, fragfunc, opts):
"""
Generates an image of a molecule depicting the fragment contribution of XLogP.
:type image: oedepict.OEImageBase
:type mol: oechem.OEMolBase
:type fragfunc: function()
:type opts: oedepict.OE2DMolDiplayOptions
"""
# calculate atom contributions of XLogP
sdatatag = "XLogP"
idatatag = oechem.OEGetTag(sdatatag)
set_atom_properties(mol, idatatag)
# fragment molecule
sgrouptag = "fragment"
igrouptag = oechem.OEGetTag(sgrouptag)
fragment_molecule(mol, fragfunc, igrouptag)
# calculate fragment contributions
minvalue, maxvalue = set_fragment_properties(mol, idatatag, igrouptag)
# initialize color gradient
lightgrey = oechem.OEColor(240, 240, 240)
colorg = oechem.OELinearColorGradient(oechem.OEColorStop(0.0, lightgrey))
colorg.AddStop(oechem.OEColorStop(minvalue, oechem.OEDarkGreen))
colorg.AddStop(oechem.OEColorStop(maxvalue, oechem.OEDarkPurple))
# generate image frames
iwidth, iheight = image.GetWidth(), image.GetHeight()
mframe = oedepict.OEImageFrame(image, iwidth, iheight * 0.8,
oedepict.OE2DPoint(0.0, 0.0))
cframe = oedepict.OEImageFrame(image, iwidth, iheight * 0.2,
oedepict.OE2DPoint(0.0, iheight * 0.8))
# initialize molecule display
opts.SetDimensions(mframe.GetWidth(), mframe.GetHeight(), oedepict.OEScale_AutoScale)
disp = oedepict.OE2DMolDisplay(mol, opts)
# initialize highlighting style
highlight = oedepict.OEHighlightByLasso(oechem.OEWhite)
highlight.SetConsiderAtomLabelBoundingBox(True)
colorgopts = oegrapheme.OEColorGradientDisplayOptions()
for group in mol.GetGroups(oechem.OEHasGroupType(igrouptag)):
groupvalue = group.GetData(idatatag)
colorgopts.AddMarkedValue(groupvalue)
# depict fragment contribution
color = colorg.GetColorAt(groupvalue)
highlight.SetColor(color)
abset = oechem.OEAtomBondSet(group.GetAtoms(), group.GetBonds())
oedepict.OEAddHighlighting(disp, highlight, abset)
# render molecule and color gradient
oedepict.OERenderMolecule(mframe, disp)
oegrapheme.OEDrawColorGradient(cframe, colorg, colorgopts)
|
Hint
You can easily adapt this example to visualize other atom properties as fragment contributions by writing your own set_atom_properties and set_fragment_properties functions.
Usage (fragxlogp2img)¶
Usage
The following commands will generate the image shown in Figure 1.
prompt > echo "SCCNC(=O)c2ccc3c(c2)sc(n3)NC(=O)NCC" > molecule.ism
prompt > python3 fragxlogp2img.py molecule.ism fragxlogp.png
Command Line Parameters¶
Simple parameter list
-height : Height of output image
-width : Width of output image
molecule display options :
-aromstyle : Aromatic ring display style
input/output options
-in : Input molecule filename
-out : Output filename of the generated image
fragmentation options
-fragtype : Fragmentation type
Discussion¶
The example above shows how to visualize the fragment contributions for a single molecule, however you might want to visualize the XLogP data for a set of molecules. In the depict_molecules_fragment_xlogp function below, each molecule is rendered into a cell of an OEReport object. The OEReport class is a layout manager allowing generation of multi-page images in a convenient way. You can see the generated multi-page PDF in Table 1.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 | def depict_molecules_fragment_xlogp(report, mollist, fragfunc, opts):
"""
Generates a report of molecules depicting the fragment contribution of XLogP.
:type report: oedepict.OEReport
:type mollist: list[oechem.OEMolBase]
:type fragfunc: function()
:type opts: oedepict.OE2DMolDiplayOptions
"""
# calculate atom contributions of XLogP
sdatatag = "XLogP"
idatatag = oechem.OEGetTag(sdatatag)
for mol in mollist:
set_atom_properties(mol, idatatag)
# fragment molecules
sgrouptag = "fragment"
igrouptag = oechem.OEGetTag(sgrouptag)
for mol in mollist:
fragment_molecule(mol, fragfunc, igrouptag)
# calculate fragment contributions
minvalue = float("inf")
maxvalue = float("-inf")
for mol in mollist:
minvalue, maxvalue = set_fragment_properties(mol, idatatag, igrouptag,
minvalue, maxvalue)
# initialize color gradient
lightgrey = oechem.OEColor(240, 240, 240)
colorg = oechem.OELinearColorGradient(oechem.OEColorStop(0.0, lightgrey))
colorg.AddStop(oechem.OEColorStop(minvalue, oechem.OEDarkGreen))
colorg.AddStop(oechem.OEColorStop(maxvalue, oechem.OEDarkPurple))
# initialize highlighting style
highlight = oedepict.OEHighlightByLasso(oechem.OEWhite)
highlight.SetConsiderAtomLabelBoundingBox(True)
for mol in mollist:
# generate image frames
cell = report.NewCell()
cwidth, cheight = cell.GetWidth(), cell.GetHeight()
mframe = oedepict.OEImageFrame(cell, cwidth, cheight * 0.8,
oedepict.OE2DPoint(0.0, 0.0))
cframe = oedepict.OEImageFrame(cell, cwidth, cheight * 0.2,
oedepict.OE2DPoint(0.0, cheight * 0.8))
# initialize molecule display
opts.SetDimensions(mframe.GetWidth(), mframe.GetHeight(), oedepict.OEScale_AutoScale)
disp = oedepict.OE2DMolDisplay(mol, opts)
colorgopts = oegrapheme.OEColorGradientDisplayOptions()
for group in mol.GetGroups(oechem.OEHasGroupType(igrouptag)):
groupvalue = group.GetData(idatatag)
colorgopts.AddMarkedValue(groupvalue)
# depict fragment contribution
color = colorg.GetColorAt(groupvalue)
highlight.SetColor(color)
abset = oechem.OEAtomBondSet(group.GetAtoms(), group.GetBonds())
oedepict.OEAddHighlighting(disp, highlight, abset)
# render molecule and color gradient
oedepict.OERenderMolecule(mframe, disp)
oegrapheme.OEDrawColorGradient(cframe, colorg, colorgopts)
|
| page 1 | page 2 | page 3 |
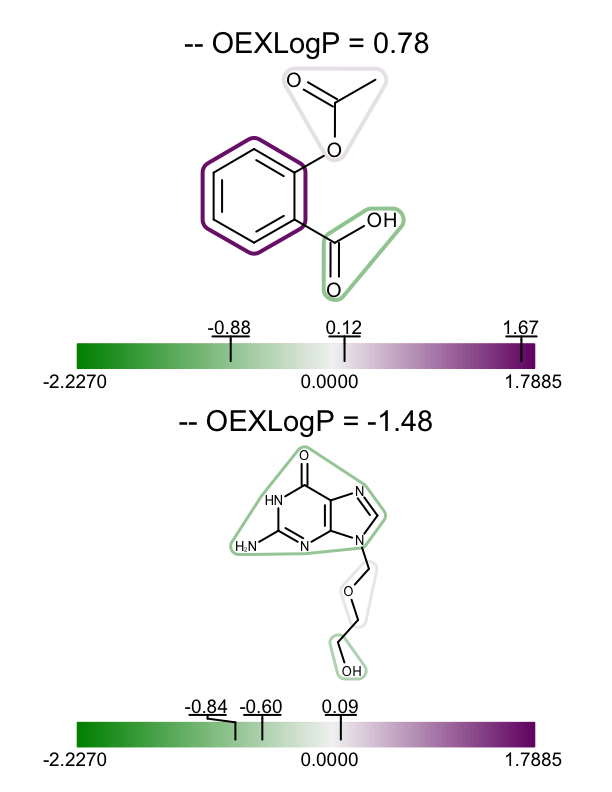
|
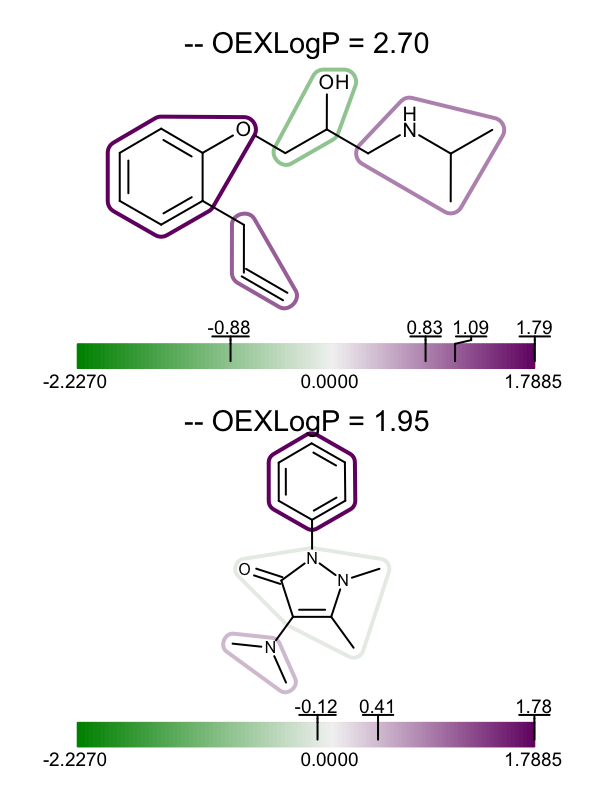
|
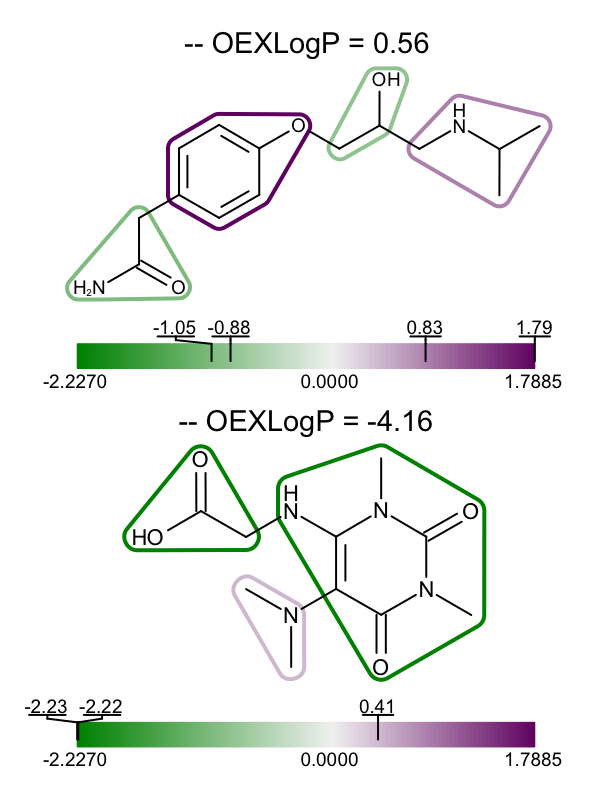
|
Usage (fragxlogp2pdf)¶
Usage
fragxlogp2pdf.py and supporting data examples.ism
The following command will generate the report shown in Table 1.
prompt > python3 fragxlogp2pdf.py examples.ism fragxlogp.pdf
Command Line Parameters¶
Simple parameter list
molecule display options :
-aromstyle : Aromatic ring display style
report options
-pagebypage : Write individual numbered separate pages
report options :
-colsperpage : Number of columns per page
-pageheight : Page height
-pageorientation : Page orientation
-pagesize : Page size
-pagewidth : Page width
-rowsperpage : Number of rows per page
input/output options
-in : Input molecule filename
-out : Output filename of the generated image
fragmentation options
-fragtype : Fragmentation type
See also in OEChem TK manual¶
Theory
- Generic Data chapter
API
- OEAtomBondSet class
- OEColorStop class
- OEGroupBase class
- OEHasGroupType functor
- OELinearColorGradient class
- OEMolBase.NewGroup method
- OEMolBase.GetGroups method
See also in OEMedChem TK manual¶
Theory
- Molecule Fragmentation chapter
API
- OEGetFuncGroupFragments function
- OEGetRingChainFragments function
- OEGetRingLinkerSideChainFragments function
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
- Highlighting chapter
- Multi Page Reports section
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEAddHighlighting function
- OEHighlightByCogwheel class
- OEImage class
- OEImageFrame class
- OERenderMolecule function
- OEReport class
