Depicting Topological Polar Surface Area¶
Problem¶
You want to depict the topological polar surface area (TPSA) of a molecule. See example in Figure 1.
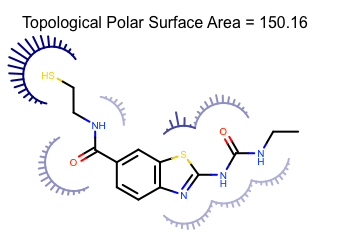
Figure 1. Example of depiction of topological polar surface area
Ingredients¶
|
Solution¶
The code snippet below shows how to calculate the total polar surface area of a molecule along with the atom contributions by calling the OEGet2dPSA function. Each atom contribution is then attached to the relevant atom as generic data with the given tag.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | def SetAtomProperties(mol, tag, minvalue=float("inf"), maxvalue=float("-inf"), SAndP=True):
avals = oechem.OEFloatArray(mol.GetMaxAtomIdx())
psa = oemolprop.OEGet2dPSA(mol, avals, SAndP)
mol.SetTitle(mol.GetTitle() + "Topological Polar Surface Area = %.2f" % psa)
for atom in mol.GetAtoms():
val = avals[atom.GetIdx()]
atom.SetData(tag, val)
minvalue = min(minvalue, val)
maxvalue = max(maxvalue, val)
return minvalue, maxvalue
|
The PSAArcFxn class below shows how to project the atom contributions of the polar surface area into a molecule surface of an atom . The __call__ method of the class takes a OESurfaceArc object that stores data required for drawing the arcs of molecule surface. The color of the arc of the molecule surface is determined by the polar surface area value attached to atom (lines 19-21). The molecule surface is rendered by using the OEDrawEyelashSurfaceArc function that draws an “eyelash” style arc with the given parameters (lines 28-36). In this case the darker colors and longer spikes indicate larger polar surface area contributions.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 | class PSAArcFxn(oegrapheme.OESurfaceArcFxnBase):
def __init__(self, colorg, tag, pen):
oegrapheme.OESurfaceArcFxnBase.__init__(self)
self.colorg = colorg
self.tag = tag
self.pen = pen
def __call__(self, image, arc):
adisp = arc.GetAtomDisplay()
if adisp is None or not adisp.IsVisible():
return False
atom = adisp.GetAtom()
atompsa = atom.GetData(self.tag)
if atompsa == 0.0:
return True
pen = oedepict.OEPen(self.pen)
color = self.colorg.GetColorAt(atompsa)
pen.SetForeColor(color)
center = arc.GetCenter()
bgnAngle = arc.GetBgnAngle()
endAngle = arc.GetEndAngle()
radius = arc.GetRadius()
edgeAngle = 10.0
patternDirection = oegrapheme.OEPatternDirection_Outside
patternAngle = 10.0
minPatternWidthRatio = 0.05
maxPatternWidthRatio = 0.70
actPatternWidthRatio = min(maxPatternWidthRatio, atompsa * (maxPatternWidthRatio / 40.0))
oegrapheme.OEDrawEyelashSurfaceArc(image, center, bgnAngle, endAngle, radius, pen,
edgeAngle, patternDirection, patternAngle,
minPatternWidthRatio, actPatternWidthRatio)
return True
def CreateCopy(self):
return PSAArcFxn(self.colorg, self.tag, self.pen).__disown__()
|
The DepictMoleculeWithPSA function below shows how to render a molecule along with its molecule surface customized to visualize the atom contributions of the polar surface area. After assigning the atom contributions to each atom (lines 6-7), an OELinearColorGradient object is constructed using the minimum and maximum contributions. This color gradient is used by the PSAArcFxn class to color the molecule surfaces based on the polar surface area values. The PSAArcFxn class that defines the style how the molecule surface is rendered is the attached to each atom by calling the OESetSurfaceArcFxn function (lines 14-15). Finally, the molecule is rendered along with its molecule surface by calling the OEDraw2DSurface function (lines 17-19).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | def DepictMoleculeWithPSA(image, mol, opts):
scale = oegrapheme.OEGetMoleculeSurfaceScale(mol, opts)
opts.SetScale(scale)
tag = oechem.OEGetTag("PSA")
minvalue, maxvalue = SetAtomProperties(mol, tag, True)
ncolor = oechem.OEColorStop(minvalue, oechem.OEWhite)
pcolor = oechem.OEColorStop(maxvalue, oechem.OEDarkBlue)
colorg = oechem.OELinearColorGradient(ncolor, pcolor)
arcfxn = PSAArcFxn(colorg, tag, opts.GetDefaultBondPen())
for atom in mol.GetAtoms():
oegrapheme.OESetSurfaceArcFxn(mol, atom, arcfxn)
disp = oedepict.OE2DMolDisplay(mol, opts)
oegrapheme.OEDraw2DSurface(disp)
oedepict.OERenderMolecule(image, disp)
|
Hint
You can easily adapt this example to visualize other atom properties by writing your own SetAtomProperties function.
Download code
Usage:
prompt > python3 psa2img.py molecule.ism psa.png
Discussion¶
The example above shows how to visualize the polar surface area for a single molecule, however you might want to visualize the polar surface area for a set of molecules.
The DepictMoleculesWithPSA function shows how the minimum and maximum polar surface area values are calculated for a set of molecules (lines 10-13). These values are then used to initialize the linear color gradient that is used to color the molecule surfaces base on the polar surface area (lines 15-19). The molecule surface is then calculated for each molecule (lines 23-24) and the molecule along with its molecule surface is rendered into a cell of an OEReport object (lines 26-30). The OEReport class is a layout manager that allows generation of multi-page images in a convenient way. You can see the generated multi-page PDF in Table 1.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | def DepictMoleculesWithPSA(report, mollist, opts):
molscale = float("inf")
for mol in mollist:
molscale = min(molscale, oegrapheme.OEGetMoleculeSurfaceScale(mol, opts))
opts.SetScale(molscale)
tag = oechem.OEGetTag("PSA")
minvalue = float("inf")
maxvalue = float("-inf")
for mol in mollist:
minvalue, maxvalue = SetAtomProperties(mol, tag, minvalue, maxvalue, True)
ncolor = oechem.OEColorStop(minvalue, oechem.OEWhite)
pcolor = oechem.OEColorStop(maxvalue, oechem.OEDarkBlue)
colorg = oechem.OELinearColorGradient(ncolor, pcolor)
arcfxn = PSAArcFxn(colorg, tag, opts.GetDefaultBondPen())
for mol in mollist:
for atom in mol.GetAtoms():
oegrapheme.OESetSurfaceArcFxn(mol, atom, arcfxn)
disp = oedepict.OE2DMolDisplay(mol, opts)
oegrapheme.OEDraw2DSurface(disp)
cell = report.NewCell()
oedepict.OERenderMolecule(cell, disp)
|
| page 1 | page 2 | page 3 |
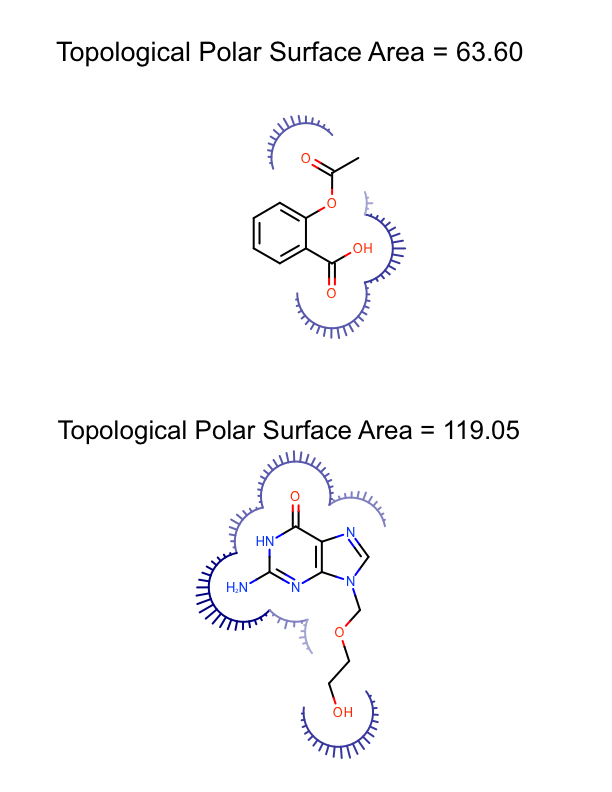
|
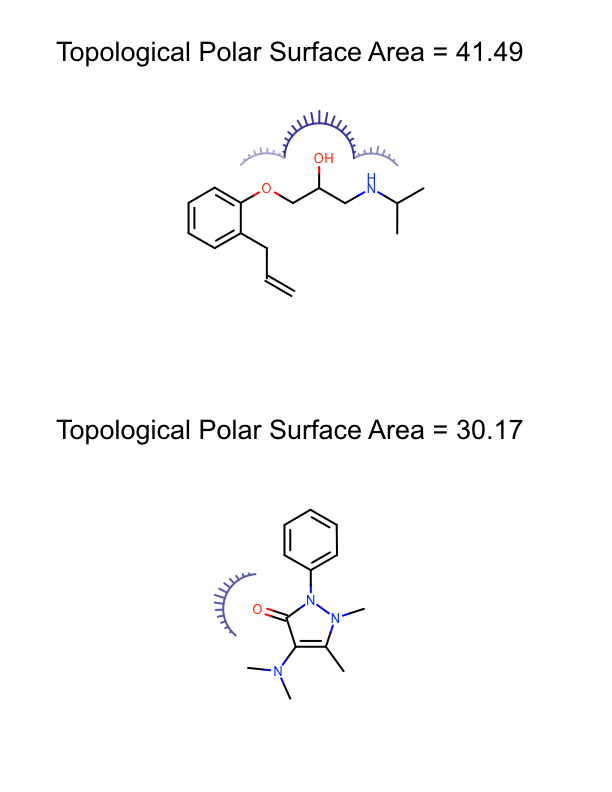
|
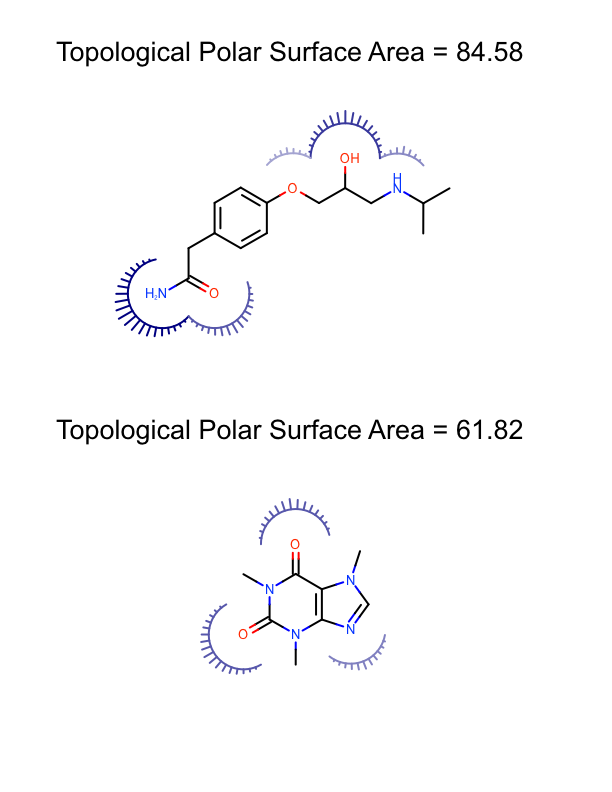
|
Download code
Usage:
prompt > python3 psa2pdf.py molecule.sdf psa.pdf
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
- Multi Page Reports section
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEImage class
- OERenderMolecule function
- OEReport class
See also in GraphemeTM TK manual¶
Theory
- Drawing a Molecule Surface chapter
API
- OEDraw2DSurface function
- OEDrawEyelashSurfaceArc function
- OEGetMoleculeSurfaceScale function
- OESetSurfaceArcFxn function
- OESurfaceArcFxnBase abstract base class
