Depicting Atom Properties¶
Problem¶
You want to depict arbitrary atom properties calculated by another application or script. See examples in Table 1..
| property map | atom glyph | molecule surface |
|
|
|
Ingredients¶
|
Download¶
Download code
atomprop2img.py and supplementary scripts: addxlogp.py addpartialcharge.py
See also the Usage (Partial Charge) and Usage (XLogP) subsections.
Solution¶
The OEChem TK provides a framework to associate arbitrary data with objects such as molecules, atoms and bonds. Generic data can be attached to an object by association either with an integer or string, called a tag identifier. When a molecule is written into an OEBinary (oeb) file, the generic data attached to the molecule and its atoms and bonds is written to the binary file as well. This provides a convenient way to transfer data along with the molecules from one application to another.
The depict_atom_property function visualizes properties attached to atom as generic data. First an image is divided into two frames, one for rendering a molecule with the atom properties and one for depicting the corresponding color gradient that shows the range of the property (lines 14-18). The scaling of the depiction options is then adjusted by utilizing the OEGetMoleculeSurfaceScale function. A OELinearColorGradient object is then initialized by calling the get_color_gradient function. This color gradient is used to associate the atom properties with colors that will represent them in the image. After constructing the molecule display, the atom properties are depicted on the molecular graph based on the user defined depiction style (lines 27-34). The molecule display and the color gradient are then rendered into the image frames along with drawing the label of the atom property into the image (lines 36-41).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 | def depict_atom_property(image, mol, opts, tagname, ncolor, pcolor, style):
"""
Depicts atom property using various styles.
:type image: oedepict.OEImageBase
:type mol: oedepict.OEMolBase
:type opts: oechem.OE2DMolDisplayOptions
:type tagname: string
:type ncolor: oechem.OEColor
:type pcolor: oechem.OEColor
:type style: string
"""
mwidth, mheight = image.GetWidth(), image.GetHeight() * 0.9
cwidth, cheight = image.GetWidth(), image.GetHeight() * 0.1
mframe = oedepict.OEImageFrame(image, mwidth, mheight, oedepict.OE2DPoint(0.0, 0.0))
cframe = oedepict.OEImageFrame(image, cwidth, cheight, oedepict.OE2DPoint(0.0, mheight))
opts.SetDimensions(mwidth, mheight, oedepict.OEScale_AutoScale)
opts.SetScale(oegrapheme.OEGetMoleculeSurfaceScale(mol, opts))
colorg = get_color_gradient(mol, tagname, ncolor, pcolor)
disp = oedepict.OE2DMolDisplay(mol, opts)
if style == "propmap":
depict_atom_property_propmap(disp, tagname, ncolor, pcolor)
if style == "atomglyph":
depict_atom_property_atomglyph(disp, tagname, colorg)
if style == "molsurface":
depict_atom_property_molsurface(disp, tagname, colorg)
oedepict.OERenderMolecule(mframe, disp)
oegrapheme.OEDrawColorGradient(cframe, colorg)
font = oedepict.OEFont(oedepict.OEFontFamily_Default, oedepict.OEFontStyle_Default, 14,
oedepict.OEAlignment_Left, oechem.OEBlack)
cframe.DrawText(oedepict.OE2DPoint(10.0, -10.0), tagname, font)
|
The get_min_max_atom_property function loops over the atoms of the molecule to find the minimum and maximum values attached to the atoms by the given tag identifier.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | def get_min_max_atom_property(mol, tagname):
"""
Calculates minimum and maximum atom property values.
:type mol: oedepict.OEMolBase
:type tagname: string
:rtype: (int, int)
"""
minvalue = float("inf")
maxvalue = float("-inf")
tag = oechem.OEGetTag(tagname)
for atom in mol.GetAtoms():
if atom.HasData(tag):
val = atom.GetData(tag)
minvalue = min(minvalue, val)
maxvalue = max(maxvalue, val)
return minvalue, maxvalue
|
The get_color_gradient function creates a OELinearColorGradient object based on the minimum and maximum atom property values.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 | def get_color_gradient(mol, tagname, ncolor, pcolor):
"""
Generates color gradient.
:type mol: oechem.OEMolBase
:type tagname: string
:type ncolor: oechem.OEColor
:type pcolor: oechem.OEColor
:rtype: oechem.OELineraColorGradient
"""
minvalue, maxvalue = get_min_max_atom_property(mol, tagname)
colorg = oechem.OELinearColorGradient(oechem.OEColorStop(0.0, oechem.OEWhite))
if minvalue < 0.0:
colorg.AddStop(oechem.OEColorStop(minvalue, ncolor))
if maxvalue > 0.0:
colorg.AddStop(oechem.OEColorStop(maxvalue, pcolor))
return colorg
|
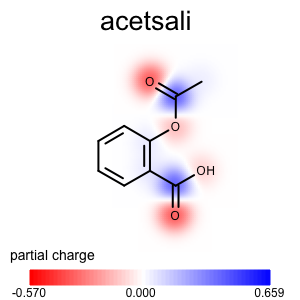
The depict_atom_property_propmap function shows how to project atom properties into a property map.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | def depict_atom_property_propmap(disp, tagname, ncolor, pcolor):
"""
Depicts atom property using property map style.
:type disp: oedepict.OE2DMolDisplay
:type tagname: string
:type ncolor: oechem.OEColor
:type pcolor: oechem.OEColor
"""
opts = disp.GetOptions()
propmap = oegrapheme.OE2DPropMap(opts.GetBackgroundColor())
propmap.SetLegendLocation(oegrapheme.OELegendLocation_Hidden)
propmap.SetNegativeColor(ncolor)
propmap.SetPositiveColor(pcolor)
propmap.Render(disp, tagname)
|
See also
- OE2DPropMap class
- Depicting Atom Property Maps chapter in Grapheme TK manual
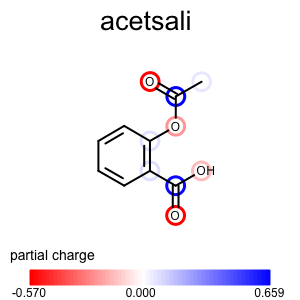
The depict_atom_property_atomglyph function shows how to visualize atom properties using atom glyphs.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 | def depict_atom_property_atomglyph(disp, tagname, colorg):
"""
Depicts atom property using atom glyph style.
:type disp: oedepict.OE2DMolDisplay
:type tagname: string
:type colorg: oechem.OELinearColorGradient
"""
tag = oechem.OEGetTag(tagname)
mol = disp.GetMolecule()
for atom in mol.GetAtoms():
if atom.HasData(tag):
value = atom.GetDoubleData(tag)
color = colorg.GetColorAt(value)
pen = oedepict.OEPen(color, color, oedepict.OEFill_Off, 3.0)
glyph = oegrapheme.OEAtomGlyphCircle(pen, oegrapheme.OECircleStyle_Default, 1.2)
oegrapheme.OEAddGlyph(disp, glyph, oechem.OEHasAtomIdx(atom.GetIdx()))
|
See also
- OEAddGlyph function
- Annotating Atoms and Bonds chapter in Grapheme TK manual
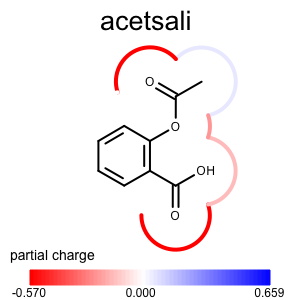
The depict_atom_property_molsurface function shows how to project atom properties into the molecule surface.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | def depict_atom_property_molsurface(disp, tagname, colorg):
"""
Depicts atom property using molecule surface style.
:type disp: oedepict.OE2DMolDisplay
:type tagname: string
:type colorg: oechem.OELinearColorGradient
"""
tag = oechem.OEGetTag(tagname)
mol = disp.GetMolecule()
for atom in mol.GetAtoms():
if atom.HasData(tag):
value = atom.GetDoubleData(tag)
color = colorg.GetColorAt(value)
pen = oedepict.OEPen(color, color, oedepict.OEFill_Off, 4.0)
oegrapheme.OESetSurfaceArcFxn(mol, atom, oegrapheme.OEDefaultArcFxn(pen))
oegrapheme.OEDraw2DSurface(disp)
|
See also
- OESetSurfaceArcFxn and OEDraw2DSurface functions
- Drawing a Molecule Surface chapter Grapheme TK manual
Discussion¶
This section shows examples to generate the input oeb file for the atomprop2img.py script.
Atom Partial Charge¶
The following example shows how to generates an OEBinary oeb file in which the partial charge is attached to each atom by the given tag string identifier using the addpartialcharge.py script.
The set_partial_charge function of the addpartialcharge.py script:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | def set_partial_charge(mol, tagname):
"""
Attaches the partial change to each atom with the given tag.
:type mol: oechem.OEMolBase
:type tagname: string
"""
oechem.OEMMFFAtomTypes(mol)
oechem.OEMMFF94PartialCharges(mol)
tag = oechem.OEGetTag(tagname)
for atom in mol.GetAtoms():
atom.SetData(tag, atom.GetPartialCharge())
|
After running the addpartialcharge.py script, the atom partial charges can be visualized by calling the atomprop2img.py script with the same tag identifier.
Usage (Partial Charge)¶
Usage
atomprop2img.py and addpartialcharge.py
Running the above commands will generate the images shown in Table 1.
prompt > echo "input.ism CC(=O)Oc1ccccc1C(=O)O acetsali" > molecule.ism
prompt > python3 addpartialcharge.py molecule.ism molecule.oeb partialcharge
prompt > python3 atomprop2img.py -in molecule.oeb -out partialcharge-propmap.png -tagname partialcharge -style propmap
prompt > python3 atomprop2img.py -in molecule.oeb -out partialcharge-atomglyph.png -tagname partialcharge -style atomglyph
prompt > python3 atomprop2img.py -in molecule.oeb -out partialcharge-molsurface.png -tagname partialcharge -style molsurface
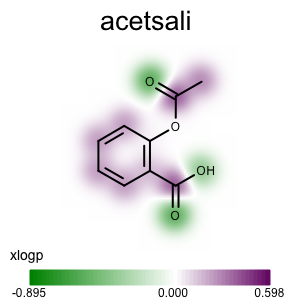
Atom XLogP Contribution¶
The following program generates an OEBinary oeb file in which the contribution of XLogP is attached to each atom by the given tag string identifier using the addxlogp.py script.
The addxlogp function of the addxlogp.py script:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | def set_xlogp(mol, tagname):
"""
Attaches the XLogP atom contribution to each atom with the given tag.
:type mol: oechem.OEMolBase
:type tagname: string
"""
oequacpac.OERemoveFormalCharge(mol)
tag = oechem.OEGetTag(tagname)
avals = oechem.OEFloatArray(mol.GetMaxAtomIdx())
oemolprop.OEGetXLogP(mol, avals)
for atom in mol.GetAtoms():
val = avals[atom.GetIdx()]
atom.SetData(tag, val)
|
After running the addxlogp.py script, the atom contributions of XLogP can be visualized by calling the atomprop2img.py script with the same tag identifier.
Usage (XLogP)¶
Usage
atomprop2img.py and addxlogp.py
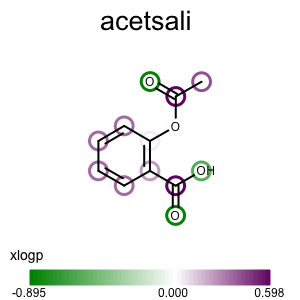
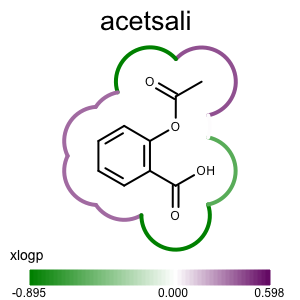
Running the above commands will generate the images shown in Table 2.
prompt > echo "input.ism CC(=O)Oc1ccccc1C(=O)O acetsali" > molecule.ism
prompt > python3 addpartialcharge.py molecule.ism molecule.oeb partialcharge
prompt > python3 atomprop2img.py -in molecule.oeb -out xlogp-propmap.png -tagname xlogp -style propmap -negcolor darkgreen -poscolor darkpurple
prompt > python3 atomprop2img.py -in molecule.oeb -out xlogp-atomglyph.png -tagname xlogp -style atomglyph -negcolor darkgreen -poscolor darkpurple
prompt > python3 atomprop2img.py -in molecule.oeb -out xlogp-molsurface.png -tagname xlogp -style molsurface -negcolor darkgreen -poscolor darkpurple
| property map | atom glyph | molecule surface |
|
|
|
See also
See also in OEChem TK manual¶
Theory
- Generic Data chapter
API
- OEColor class
- OEColorStop class
- OEGetTag function
- OELinearColorGradient class
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEFont class
- OEImage class
- OEImageFrame class
- OERenderMolecule function
See also in GraphemeTM TK manual¶
Theory
- Depicting Atom Property Maps chapter
- Annotating Atoms and Bonds chapter
- Drawing a Molecule Surface chapter
API
- OE2DPropMap class
- OEAddGlyph function
- OEAtomGlyphCircle class
- OEGetMoleculeSurfaceScale function
- OEDraw2DSurface function
- OEDrawColorGradient function
