Depicting Reaction Components¶
Problem¶
You want to depict your MDL reaction with highlighting of its reactant and product components.
OEDepict TK provides ability to depict the mapping index between reactant and product atoms. See example in Figure 1.
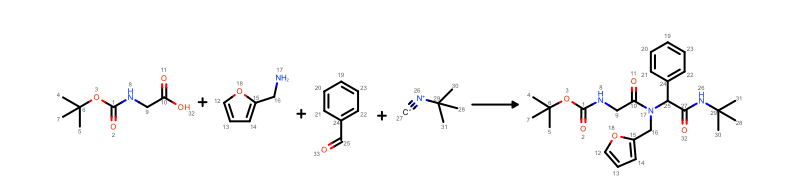
Figure 1. Example of MDL reaction depiction with atom mapping
However, there is a better way to visualize this information using highlighting. In the image below, each reactant of the reaction is highlighted using a different color and then the same color is used to highlight the atoms of the product(s) which are mapped to atoms of the given reactant. See example in Figure 2.
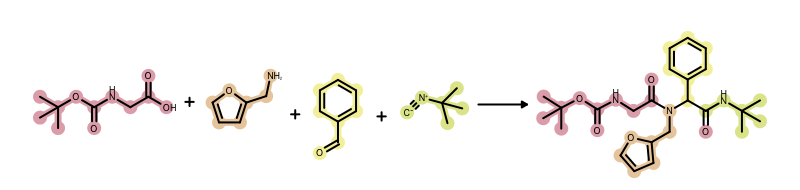
Figure 2. Example of MDL reaction depiction with component highlighting
Ingredients¶
|
Solution¶
The ColorReactionByComponents shows how a reaction is partitioned into components by calling the OEDetermineComponents function. The OEDetermineComponents function returns the number of components of a molecule i.e. in this case the number of reactants and products of the reaction and it also returns a list that stores which part each atom belongs to (line 4) . This parts list is then used to set up a OEPartPredAtom predicate (line 5). A loop over the reactant components of the reaction (line 9), associates each with different color. Inside the loop the atoms of each reactant are highlighted (lines 15-17) along with the product atoms that are mapped to the given reactant (lines 19-21).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | def ColorReactionByComponents(rdisp):
rmol = rdisp.GetMolecule()
nrparts, parts = oechem.OEDetermineComponents(rmol)
partpred = oechem.OEPartPredAtom(parts)
colors = [c for c in oechem.OEGetLightColors()]
highlightstyle = oedepict.OEHighlightStyle_BallAndStick
for partidx, color in zip(range(1, nrparts + 1), colors):
partpred.SelectPart(partidx)
if IsReactantComponent(rmol, partpred):
reactant = oechem.OEAtomBondSet(rmol.GetAtoms(partpred))
AddBondsBetweenAtoms(rmol, reactant)
oedepict.OEAddHighlighting(rdisp, color, highlightstyle, reactant)
product = GetProductAtoms(rmol, reactant)
AddBondsBetweenAtoms(rmol, product)
oedepict.OEAddHighlighting(rdisp, color, highlightstyle, product)
|
The IsReactantComponent function identifies whether a given component is a reactant in which case it returns True.
1 2 3 4 | def IsReactantComponent(rmol, partpred):
atom = rmol.GetAtom(oechem.OEAndAtom(partpred, oechem.OEAtomIsInReactant()))
return (atom is not None)
|
The GetProductAtoms function is used to identify the product atoms of the reaction that are mapped to the atoms of the given reactant. Two atoms are mapped to each other if they have the same index returned by the OEAtomBase.GetMapIdx method.
1 2 3 4 5 6 7 8 9 10 11 12 | def GetProductAtoms(rmol, reactant):
product = oechem.OEAtomBondSet()
for ratom in reactant.GetAtoms():
mapidx = ratom.GetMapIdx()
if mapidx != 0:
patom = rmol.GetAtom(oechem.OEAndAtom(oechem.OEHasMapIdx(mapidx),
oechem.OEAtomIsInProduct()))
if patom is not None:
product.AddAtom(patom)
return product
|
In order to highlight not only the atoms but also the bonds of the reaction the AddBondsBetweenAtoms function is called. This adds a bond to the given OEAtomBondSet object if both of its end atoms belong to the same component.
1 2 3 4 5 6 | def AddBondsBetweenAtoms(rmol, abset):
pred = oechem.OEIsAtomMember(abset.GetAtoms())
for bond in rmol.GetBonds():
if pred(bond.GetBgn()) and pred(bond.GetEnd()):
abset.AddBond(bond)
|
Download code
reaction2img.py and supporting data set ugi.rxn
Usage:
prompt > python3 reaction2img.py ugi.rxn ugi.png
See also¶
- Ugi reaction in Wikipedia
See also in OEChem TK manual¶
Theory
- Predicate Functors chapter
- MDL Reaction Query File section
API
- OEAtomBase.GetMapIdx method
- OEAtomIsInReactant predicate
- OEAtomIsInProduct predicate
- OEAtomBondSet class
- OEDetermineComponents function
- OEHasMapIdx predicate
- OEIsAtomMember predicate
- OEPartPredAtom predicate
See also in OEDepict TK manual¶
Theory
- Highlighting chapter
- MDL Reaction Depiction chapter
API
- OEAddHighlighting function
