Depicting Polar Hydrogens¶
Problem¶
You want to depict your molecule with explicit polar hydrogens. See example in Figure 1.
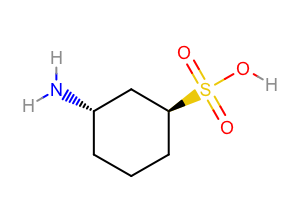
Figure 1. Example of depicting polar hydrogens
Ingredients¶
|
Difficulty Level¶

Solution¶
Before rendering a molecule, the OEPrepareDepiction function should be called which prepares the molecule for depiction. This function perceives atom and bond stereo information and suppresses or adds hydrogens if necessary. By default, the explicit hydrogens are suppressed by the OEPrepareDepiction function, see example (A) in Table: Example of default molecule depiction. Only hydrogens that are necessary to faithfully represent tetrahedral stereochemistry are added, see example (B) in Table 1.
| ( A ) | ( B ) |
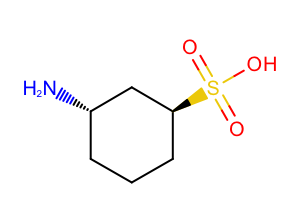
|
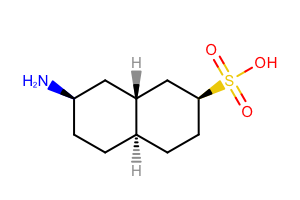
|
If the molecule has only implicit hydrogens then you can add the polar hydrogens by calling the OEAddExplicitHydrogens function with the polarOnly = True parameter (lines 3-4), before preparing the molecule for depiction. It is important to call the OEPrepareDepiction function with clearcoords = True and suppressH = False parameters (lines 6-8).
1 2 3 4 5 6 7 8 9 10 11 | def DepictMoleculeWithPolarHydrogens(mol, opts, filename):
polarOnly = True
oechem.OEAddExplicitHydrogens(mol, polarOnly)
clearcoords = True
suppressH = False
oedepict.OEPrepareDepiction(mol, clearcoords, suppressH)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule(filename, disp)
|
Discussion¶
If the molecule already has explicit hydrogens then you can suppress all non polar hydrogens by calling the OESuppressHydrogens function with retainPolar = True parameter (lines 3-4), prior to preparing the molecule for depiction.
1 2 3 4 5 6 7 8 9 10 11 | def DepictMoleculeOnlyPolarHydrogens(mol, opts, filename):
retainPolar = True
oechem.OESuppressHydrogens(mol, retainPolar)
clearcoords = True
suppressH = False
oedepict.OEPrepareDepiction(mol, clearcoords, suppressH)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule(filename, disp)
|
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEAddDepictionHydrogens function
- OEPrepareDepiction function
- OERenderMolecule function