Depicting Molecule with Various Styles¶
Problem¶
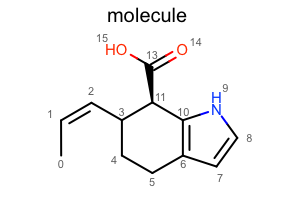
You want to depict a molecule with various styles.
|
|
|
|
|
|
|
|
|
|
|
|
The examples below generate interactive .svg images These svg images should be included into and HTML page with the SVG MIME type.
<object data="<imagename>.svg" type="image/svg+xml"></object>
|
|
|
|
Ingredients¶
|
Difficulty Level¶

Solution¶
Default¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-default.png", disp)
Download code |
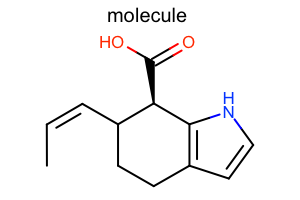
|
See also
- OE2DMolDisplayOptions class
- OE2DMolDisplay class
- OERenderMolecule function
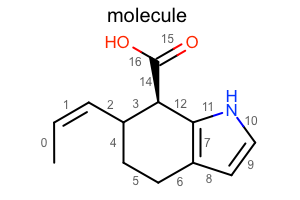
Aromaticity Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAromaticStyle(oedepict.OEAromaticStyle_Circle)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-aromatic-style.png", disp)
Download code |
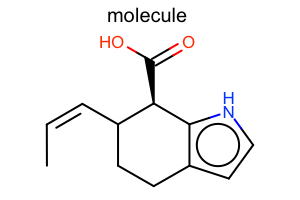
|
See also
- OE2DMolDisplayOptions.SetAromaticStyle method
- OEAromaticStyle namespace
Atom Color Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomColorStyle(oedepict.OEAtomColorStyle_BlackCPK)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-color-style.png", disp)
Download code |
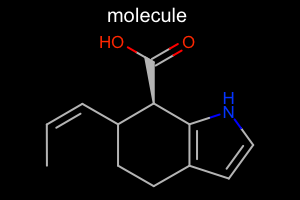
|
See also
- OE2DMolDisplayOptions.SetAtomColorStyle method
- OEAtomColorStyle namespace
Super Atom Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetSuperAtomStyle(oedepict.OESuperAtomStyle_All)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-super-atom-style.png", disp)
Download code |
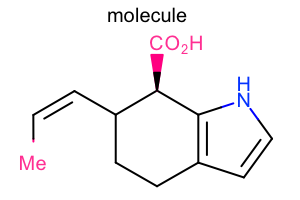
|
See also
- OE2DMolDisplayOptions.SetSuperAtomStyle method
- OESuperAtomStyle namespace
Atom Stereo Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
stereostyle = oedepict.OEAtomStereoStyle_Display_All
stereostyle |= oedepict.OEAtomStereoStyle_HashWedgeStyle_Standard
opts.SetAtomStereoStyle(stereostyle)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-stereo-style.png", disp)
Download code |
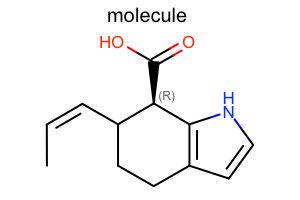
|
See also
- OE2DMolDisplayOptions.SetAtomStereoStyle method
- OEAtomStereoStyle namespace
Bond Stereo Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondStereoStyle(oedepict.OEBondStereoStyle_Display_All)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-stereo-style.png", disp)
Download code |
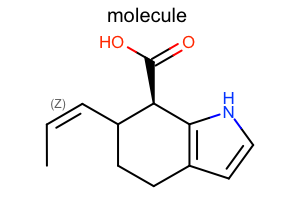
|
See also
- OE2DMolDisplayOptions.SetBondStereoStyle method
- OEBondStereoStyle namespace
Title Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetTitleLocation(oedepict.OETitleLocation_Bottom)
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold | oedepict.OEFontStyle_Italic
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OERoyalBlue)
opts.SetTitleFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-title.png", disp)
Download code |
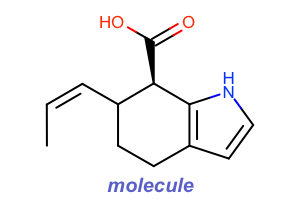
|
See also
- OE2DMolDisplayOptions.SetTitleLocation method
- OETitleLocation namespace
- OEFont class
- OE2DMolDisplayOptions.SetTitleFont method
Border Style¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
disp = oedepict.OE2DMolDisplay(mol, opts)
layer = disp.GetLayer(oedepict.OELayerPosition_Below)
lightred = oechem.OEColor(255, 235, 235) # R, G, B
mediumred = oechem.OEColor(225, 55, 55) # R, G, B
linewidth = 6.0
pen = oedepict.OEPen(lightred, mediumred, oedepict.OEFill_On, linewidth)
cornersize = 30
oedepict.OEDrawCurvedBorder(layer, pen, cornersize)
oedepict.OERenderMolecule("depiction-border.png", disp)
Download code |
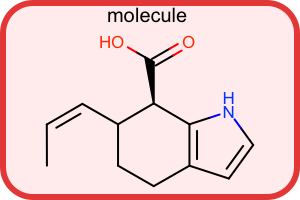
|
See also
- OE2DMolDisplay.GetLayer method
- OELayerPosition namespace
- OEPen class
- OEDrawCurvedBorder method
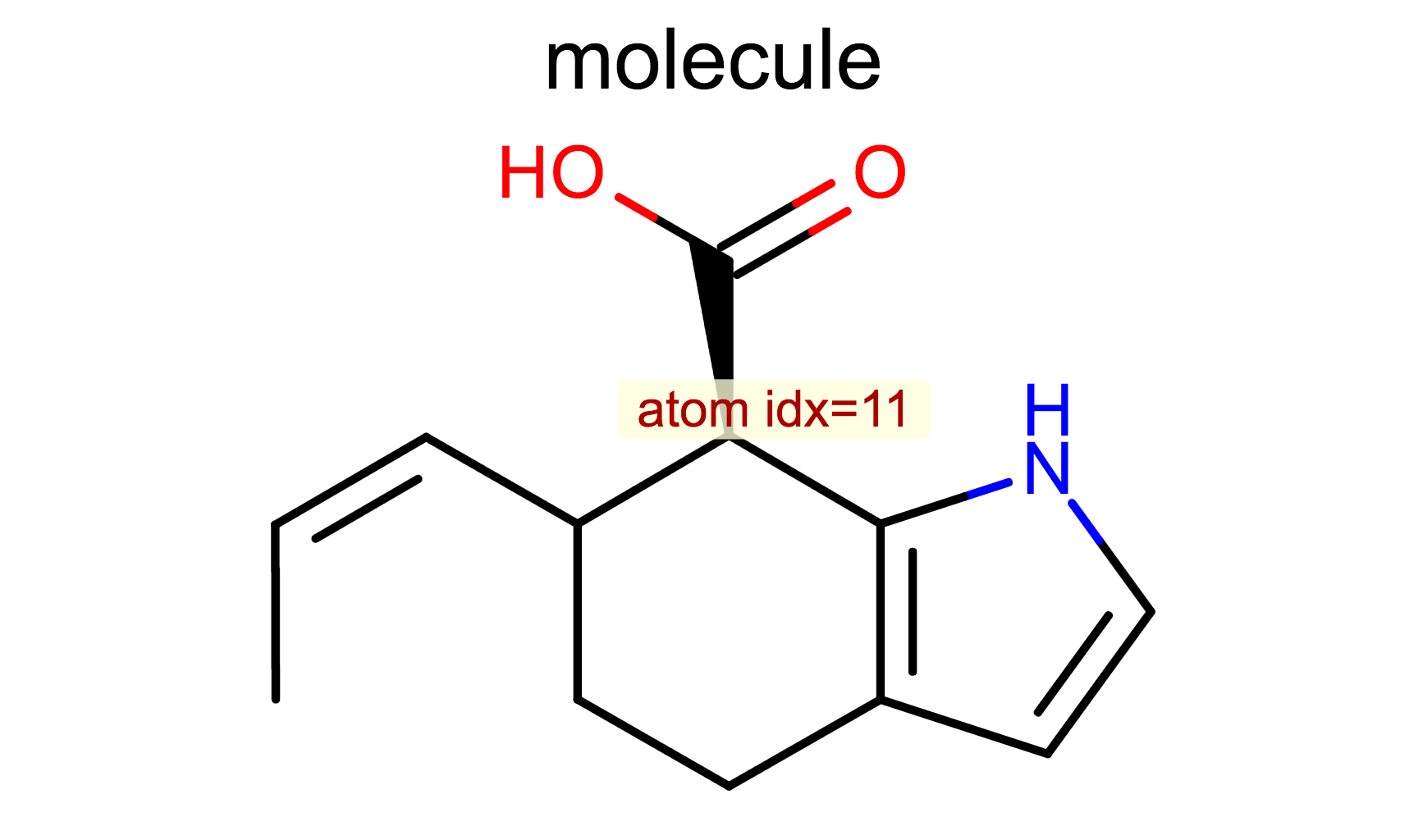
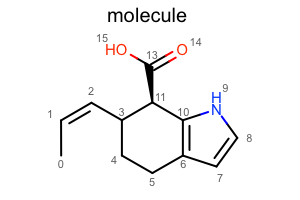
Display Atom Index¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomPropertyFunctor(oedepict.OEDisplayAtomIdx())
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-idx.png", disp)
Download code |
|
See also
- OE2DMolDisplayOptions.SetAtomPropertyFunctor method
- OEDisplayAtomIdx class
- OEDisplayAtomPropBase base class
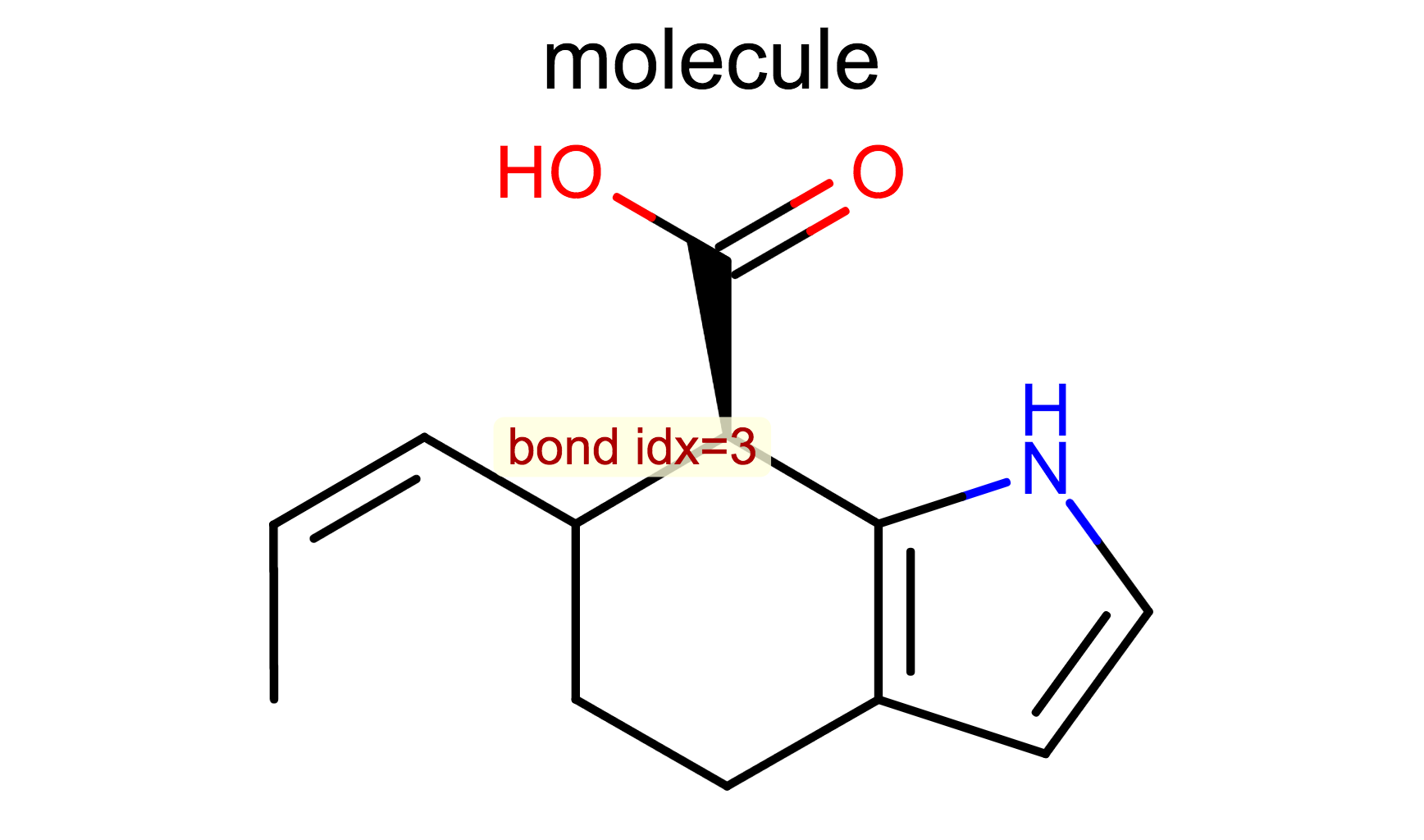
Display Bond Index¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondPropertyFunctor(oedepict.OEDisplayBondIdx())
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-idx.png", disp)
Download code |

|
See also
- OE2DMolDisplayOptions.SetBondPropertyFunctor method
- OEDisplayBondIdx class
- OEDisplayBondPropBase base class
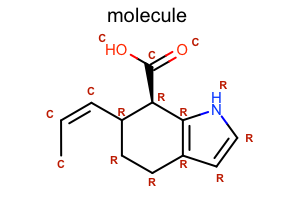
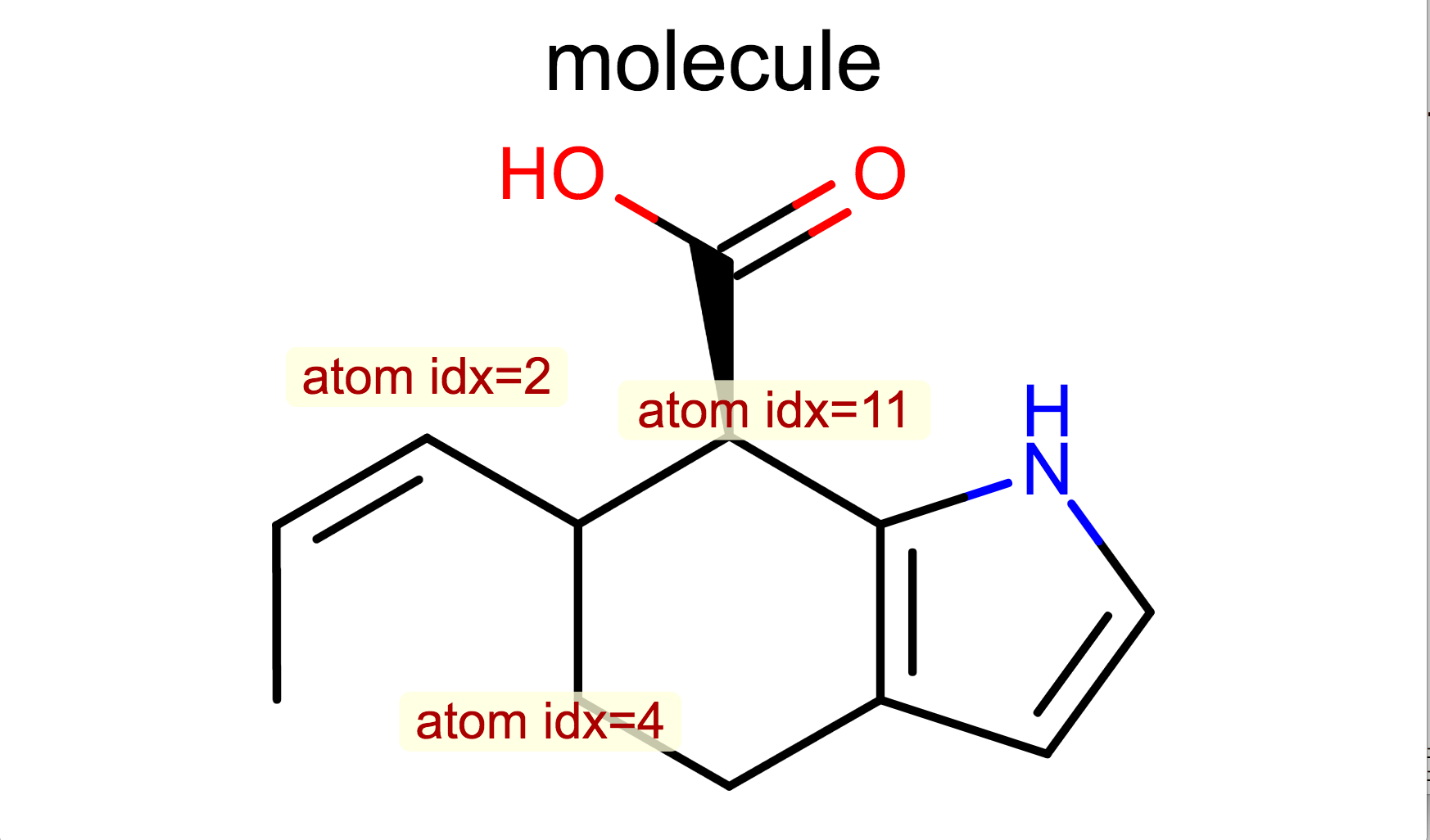
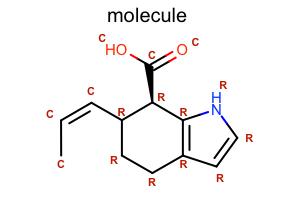
Display Atom Property¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
class LabelAtomTopology(oedepict.OEDisplayAtomPropBase):
def __init__(self):
oedepict.OEDisplayAtomPropBase.__init__(self)
def __call__(self, atom):
if atom.IsInRing():
return "R"
return "C"
def CreateCopy(self):
copy = LabelAtomTopology()
return copy.__disown__()
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetAtomPropertyFunctor(LabelAtomTopology())
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OEDarkRed)
opts.SetAtomPropLabelFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-atom-prop.png", disp)
Download code |
|
See also
- OEDisplayAtomPropBase base class
- OE2DMolDisplayOptions.SetAtomPropertyFunctor method
- OEFont class
- OE2DMolDisplayOptions.SetAtomPropLabelFont method
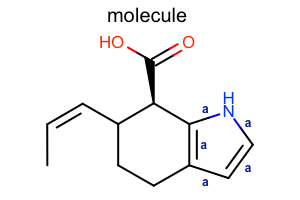
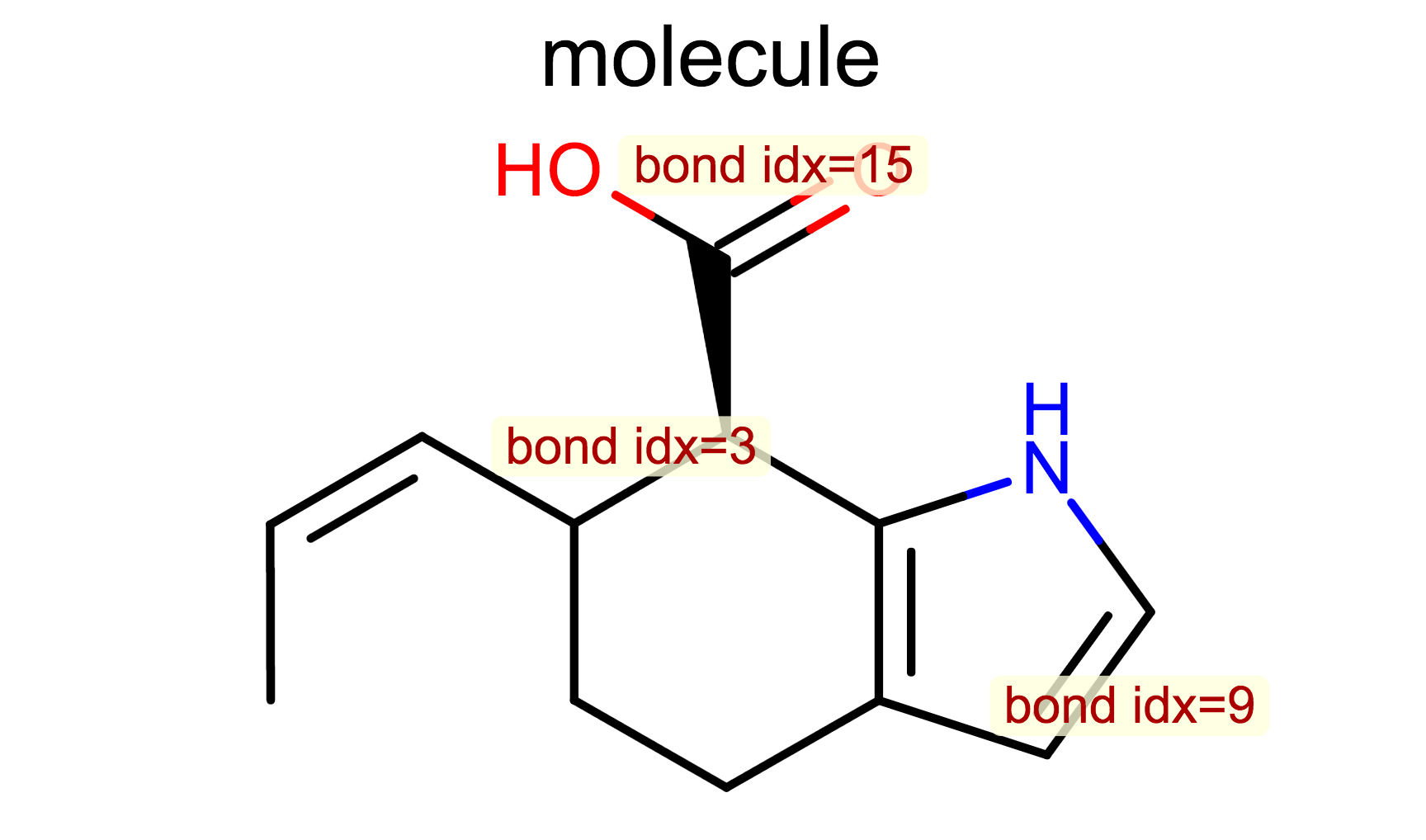
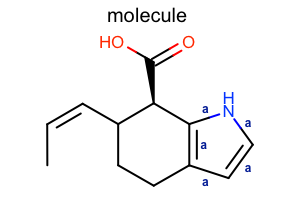
Display Bond Property¶
#!/usr/bin/env python3
from openeye import oechem
from openeye import oedepict
class LabelBondTopology(oedepict.OEDisplayBondPropBase):
def __init__(self):
oedepict.OEDisplayBondPropBase.__init__(self)
def __call__(self, bond):
if bond.IsAromatic():
return "a"
return ""
def CreateCopy(self):
copy = LabelBondTopology()
return copy.__disown__()
mol = oechem.OEGraphMol()
oechem.OESmilesToMol(mol, "C/C=C\C1CCc2cc[nH]c2[C@@H]1C(=O)O molecule")
oedepict.OEPrepareDepiction(mol)
width, height, scale = 300, 200, oedepict.OEScale_AutoScale
opts = oedepict.OE2DMolDisplayOptions(width, height, scale)
opts.SetBondPropertyFunctor(LabelBondTopology())
fonttype = oedepict.OEFontFamily_Default
fontstyle = oedepict.OEFontStyle_Bold
fontalign = oedepict.OEAlignment_Center
fontsize = 10
font = oedepict.OEFont(fonttype, fontstyle, fontsize, fontalign, oechem.OEDarkBlue)
opts.SetBondPropLabelFont(font)
disp = oedepict.OE2DMolDisplay(mol, opts)
oedepict.OERenderMolecule("depiction-bond-prop.png", disp)
Download code |
|
See also
- OEDisplayBondPropBase base class
- OE2DMolDisplayOptions.SetBondPropertyFunctor method
- OEFont class
- OE2DMolDisplayOptions.SetBondPropLabelFont method
Hover Atom Property¶
See also
- OEFont class
- OEDrawSVGHoverText function
Hover Bond Property¶
See also
- OEFont class
- OEDrawSVGHoverText function
Toggle Atom Property¶
See also
- OEFont class
- OEDrawSVGToggleText function
Toggle Bond Property¶
See also
- OEFont class
- OEDrawSVGToggleText function
See also in OEDepict TK manual¶
Theory
- Molecule Depiction chapter
- Generating Interactive SVG images chapter
API
- OE2DMolDisplay class
- OE2DMolDisplayOptions class
- OEPrepareDepiction function
- OERenderMolecule function