Aligning Molecules Based on Substructure Search
A program that aligns the molecules based on a substructure search initialized with the given SMARTS pattern.
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> smartsalign2D --help
Will generate the following output:
Simple parameter list
input/output options :
-in : Filename of input molecules
-out : Output filename
-smarts : SMARTS for alignment match
Code
Download code
smartsalign2D.cpp
and
smartsalign2D.txt
interface file
See also
Molecule Alignment chapter
OESubSearch class
OEGenerate2DCoordinatesfunctionOEIs2DFormatfunctionOEPrepareSearchfunctionOEPrepareAlignedDepictionfunctionOEPrepareDepictionfunction
Examples
prompt> smartsalign2D -in test.sdf -out aligned.sdf -smarts "c1cc[c,n]c2c1ncc2"
Taking the output coordinates and generating the images with the mol2img program will produce the images shown in Table: Example of using the program to align molecule
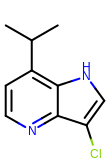
|
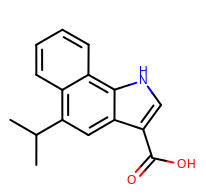
|
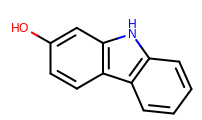
|
See also
Molecule Alignment chapter