OEGetFuncGroupFragments
OESystem::OEIterBase<OEChem::OEAtomBondSet> *
OEGetFuncGroupFragments(const OEChem::OEMolBase &mol)
The OEGetFuncGroupFragments function partitions
the given molecule into functional groups and returns an iterator
over OEAtomBondSet objects.
Each OEAtomBondSet container stores the atoms and the
bonds of an identified fragment.
See example in Figure: Example of fragments returned by the
OEGetFuncGroupFragments function where
each fragment returned by the OEGetFuncGroupFragments
function is highlighted with a different color.
The OEGetFuncGroupFragments function uses the following
heuristics to fragment a molecule:
rings are left intact i.e. considered as a unit
exo-ring double bonds are not cleaved from adjacent rings
hetero atoms next to each other are kept together
sp and sp2 atoms next to each other are kept together
in order to avoid generating one atom fragments isolated atoms are attached to the smallest neighbor fragment
Note
The molecule argument is const, so this function requires that the
OEFindRingAtomsAndBonds perception activity has been
performed prior to calling this function.
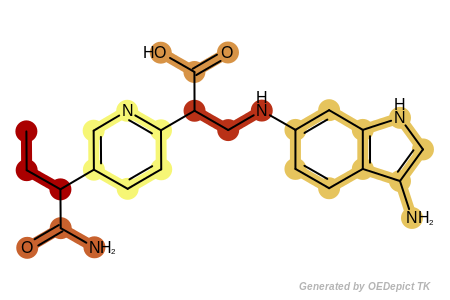
Example of fragments returned by the OEGetFuncGroupFragments function
See also
OEAtomBondSet and OEMolBase classes in the OEChem TK manual
Molecule Fragmentation chapter