OEGetRingChainFragments
OESystem::OEIterBase<OEChem::OEAtomBondSet> *
OEGetRingChainFragments(const OEChem::OEMolBase &mol)
The OEGetRingChainFragments function partitions
the given molecule into ring and chain fragments and returns an iterator
over OEAtomBondSet objects.
Each OEAtomBondSet container stores the atoms and the
bonds of an identified fragment.
See example in Figure: Example of fragments returned by the
OEGetRingChainFragments function where
each fragment returned by the OEGetRingChainFragments
function is highlighted with a different color.
The perceived regions are based upon the heavy atoms in the molecule. If the molecule contains explicit hydrogen atoms, those explicit hydrogens will be returned in the same perception set(s) as their alpha heavy atom neighbor. If the perception set(s) returned are used to subset or fragment the molecule, a mixture of implicit and explicit hydrogens may result, so care should be taken to normalize the structure for subsequent use.
Note
The molecule argument is const, so this function requires that the
OEFindRingAtomsAndBonds perception activity has been
performed prior to calling this function.
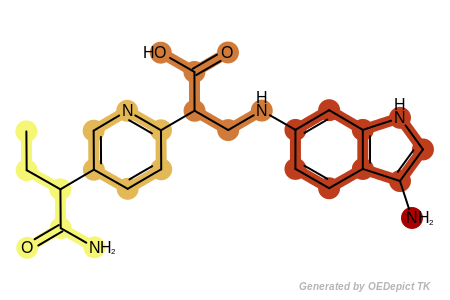
Example of fragments returned by the OEGetRingChainFragments function
See also
OEAtomBondSet and OEMolBase classes in the OEChem TK manual
Molecule Fragmentation chapter