OEGetXLogP
bool OEGetXLogP(const OEChem::OEMolBase &mol, float &xlogp, float *atomxlogps=0)
Returns the XLogP for a given molecule as described in the
LogP section. The returned value will be equal to
the sum on the individual atom contributions plus the linear
regression constant -0.127.
The atomxlogps parameter can be used to retrieve the contribution
of each atom to the total XLogP as shown in
Listing 2.
See example in Figure: Example of depicting the atom contributions
of the XLogP.
Listing 2: Example of retrieving individual atom contributions to XLogP
// declare an array for atom values
float * atomXLogP = new float[mol.GetMaxAtomIdx()];
float xlogp;
bool rc = OEGetXLogP(mol, xlogp, atomXLogP);
if (!rc)
std::cout << "XLogP failed for molecule" << std::endl;
else
{
std::cout << "XLogP = " << xlogp << std::endl;
for (OEIter<OEAtomBase> atom=mol.GetAtoms();atom;++atom)
{
unsigned int idx = atom->GetIdx();
std::cout << idx << " " << atomXLogP[idx] << std::endl;
}
}
// clean up
delete [] atomXLogP;
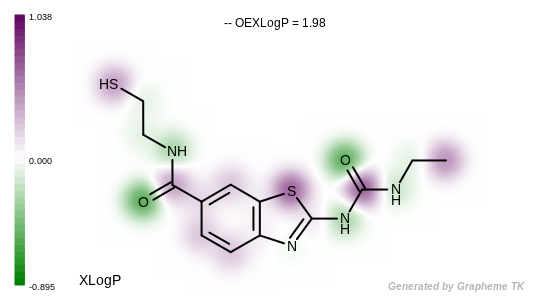
Example of depicting the atom contributions of the XLogP
Hint
XLogP should be calculated on the neutral form of the molecule.
Therefore, calling the OERemoveFormalCharge function
of Quacpac TK is highly recommended prior to the XLogP calculation.
See also
OEGetXLogPResult function
LogP section