Multi Page Images
Multi-page images can be generated by the usage of the
OEMultiPageImageFile class.
After generating a multi-page object by specifying the orientation and
the size of its pages, the individual pages can be accessed by calling
the OEMultiPageImageFile::NewPage method.
The returned OEImage then can be used to depict
molecule(s) as demonstrated in the previous chapters.
The image created by Listing 1 is shown in
Figure: Example multi-page depiction.
Listing 1: Example of multi-page depiction
const vector<string> smiles = { "C1CC(C)CCC1", "C1CC(O)CCC1", "C1CC(Cl)CCC1",
"C1CC(F)CCC1", "C1CC(Br)CCC1", "C1CC(N)CCC1" };
OEMultiPageImageFile multi(OEPageOrientation::Landscape, OEPageSize::US_Letter);
OE2DMolDisplayOptions opts;
const auto rows = 2u;
const auto cols = 2u;
std::unique_ptr<OEImageGrid> grid(nullptr);
OEIter<OEImageBase> cell;
for (const auto& smi : smiles)
{
if (!cell)
{ // go to next page
OEImage& image = multi.NewPage();
grid.reset(new OEImageGrid(image, rows, cols));
grid->SetCellGap(20);
grid->SetMargins(20);
cell = grid->GetCells();
}
OEGraphMol mol;
OESmilesToMol(mol, smi);
OEPrepareDepiction(mol);
opts.SetDimensions(cell->GetWidth(), cell->GetHeight(), OEScale::AutoScale);
OE2DMolDisplay disp(mol, opts);
OERenderMolecule(cell, disp);
OEDrawBorder(cell, OERedPen);
++cell;
}
OEWriteMultiPageImage("MultiPage.pdf", multi);
page 1 |
page 2 |
|---|---|
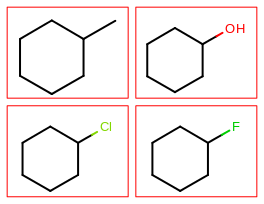
|
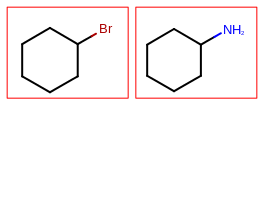
|
The Listing 1 example creates a PDF multi-page image
file, the multi-page image types supported by OEDepict TK can be accessed by
calling the OEGetSupportedMultiPageImageFileExtensions
function.
It returns an iterator over the file extensions that can be used when
writing a multi-page image file by the
OEWriteMultiPageImage function.
Multi Page Reports
The Listing 1 example illustrates how depict a set of
molecules in a multi-page PDF using the OEImageGrid
and OEMultiPageImageFile classes.
OEDepict TK also provides the OEReport class to do
this task in a more convenient way.
The OEReport class is a multi-page layout manager that
handles the page generation and the positioning of information
(such as text and molecule depiction).
In the Listing 2 example, first a
OEReportOptions object is created that stores
all properties that determine the layout of a documentation.
After generating the OEReport object a
molecule can be positioned on a cell that is returned by
the OEReport::NewCell method.
The OEReport::NewCell method creates cells
from left to right and top to bottom order on each page.
If no more cells are left on the page, then a new page is created
before returning the first cell of this new page.
After the document is generated, i.e all molecule are depicted,
the OEWriteReport function writes the multi-page
documentation into into a file with the given name.
The image created by the Listing 2 example is the
same as depicted in Figure: Example multi-page depiction.
Listing 2: Example of multi-page depiction using OEReportOptions
const vector<string> smiles = { "C1CC(C)CCC1", "C1CC(O)CCC1", "C1CC(Cl)CCC1",
"C1CC(F)CCC1", "C1CC(Br)CCC1", "C1CC(N)CCC1" };
const auto rows = 2u;
const auto cols = 2u;
OEReportOptions reportopts(rows, cols);
reportopts.SetPageOrientation(OEPageOrientation::Landscape);
reportopts.SetCellGap(20);
reportopts.SetPageMargins(20);
OEReport report(reportopts);
OE2DMolDisplayOptions opts(report.GetCellWidth(), report.GetCellHeight(), OEScale::AutoScale);
for (const auto& smi : smiles)
{
OEGraphMol mol;
OESmilesToMol(mol, smi);
OEPrepareDepiction(mol);
OEImageBase* cell = report.NewCell();
OE2DMolDisplay disp(mol, opts);
OERenderMolecule(*cell, disp);
OEDrawBorder(*cell, OERedPen);
}
OEWriteReport("MultiPageReport.pdf", report);
See also
OEReportOptions class
OEReport class