Multi-Threading
OpenEye deems only the following toolkits “thread-safe”:
OEChem TK (including oeplatform, oesystem, oechem, and oebio)
OEDepict TK
OEDocking TK
Grapheme TK
GraphSim TK
Lexichem TK
Omega TK
Warning
Absolutely no guarantee is made about the thread-safety of any other library not on the preceding list.
The most common reason to multi-thread a program is for performance. However, threading has certain overhead costs associated with it. The user should profile their application to determine the best possible way to parallelize it. There is no guarantee that multi-threading a program will make it run faster, in fact, it can make it run dramatically more slowly. Some of the examples presented in this chapter will perform more slowly than their single-threaded counterparts. They were chosen for the simplicity of demonstrating certain concepts. The user should perform timing analysis on the code in this chapter on their own systems to determine whether the methods are appropriate for them.
Input and Output Threads
OEChem TK provides a rather advanced mechanism for spawning a thread to handle I/O asynchronously. Parallelizing I/O operations can yield significant performance gains when dealing with bulky file formats like SDF and MOL2, or when dealing with files on network mounted directories. Naturally, performance gains may vary based upon the system configuration in question.
The input and output threads are encapsulated inside the
oemolthread
abstraction. oemolthreads are
designed to behave as similarly as possible to
oemolstreams. This should allow easy
drop in replacement to existing code by simply changing the type of a
variable from oemolistream to
oemolithread for input, and
oemolostream to oemolothread for
output.
For example, the MolFind program described in the
Substructure Search section can be modified to the nearly
identical program in Listing 1.
Listing 1: Using Threaded Input and Output
package openeye.docexamples.oechem;
import openeye.oechem.*;
public class ThreadedMolFind {
public static void main(String argv[]) {
OESubSearch ss = new OESubSearch(argv[0]);
oemolithread ifs = new oemolithread(argv[1]);
oemolothread ofs = new oemolothread(argv[2]);
OEGraphMol mol = new OEGraphMol();
while (oechem.OEReadMolecule(ifs, mol)) {
oechem.OEPrepareSearch(mol, ss);
if (ss.SingleMatch(mol))
oechem.OEWriteMolecule(ofs, mol);
}
ofs.close();
ifs.close();
}
}
Note
Both oemolstreams and
oemolthreads should be evaluated
for performance. oemolthreads
increase user time CPU usage to save on potentially expensive
system calls.
The difference in Listing 1 may be a little
difficult to spot for the experienced OEChem TK programmer used to
seeing oemolstream declarations,
however, the following two declarations are not
oemolstreams. They are
oemolthreads meant to behave as
closely as possible to oemolstreams.
oemolithread ifs = new oemolithread(argv[1]);
oemolothread ofs = new oemolothread(argv[2]);
However, oemolthreads could not be
made to behave exactly like
oemolstreams. The following key
differences between oemolthreads and
oemolstreams should be taken into
account:
oemolthreads do not support stream
methods: read,
seek,
size, and
tell.
These do not make much sense in a multi-threaded environment where the I/O thread can be at any arbitrary location in the file. Using the methods would be prone to race conditions.
The oemolithread default
constructor does not
default to stdin.
The default constructor of oemolistream will open the stream on
stdin. However, read operations are handled by a separate opaque I/O thread inside the oemolithread. The I/O thread uses blocking reads that do not return unless the user closesstdinof the process.oemolithreadscan still read fromstdinusing the same syntactic trick described in the Command Line Format Control section supplied toopenor theconstructorwhich takes astring.
The oemolithread does not support the
SetConfTest interface.
As will be described later, the I/O threads inside
oemolthreadsdo a minimal amount of work to maximize scalability in parallel applications. The work described in the Input and Output section to recover multi-conformer molecules from single-conformer file formats can be quite expensive. Note, this does not precludeoemolithreadsfrom reading multi-conformer molecules from file formats that support them inherently, e.g., multi-conformer molecules can be read out of multi-conformer OEB files throughoemolithreads.
With all the preceding considerations taken into account the real gain
of oemolthreads is that
OEReadMolecule and OEWriteMolecule
can be safely called from multiple threads onto the same
oemolthread. They are guaranteed to
be free of race conditions when applied to
oemolthreads. Not only that, the
expensive parsing (OEReadMolecule) and serialization
(OEWriteMolecule) occurs in the thread that calls
the function, not in the oemolthread
itself. The oemolthread takes care
of I/O and compression if reading or writing to a .gz file.
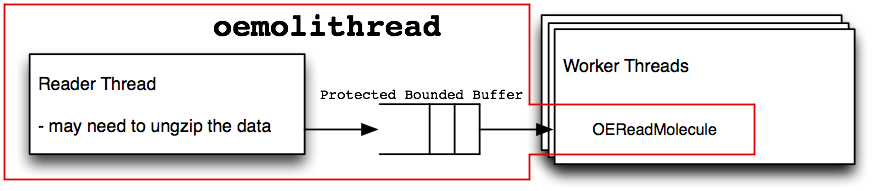
oemolithread
Diagram of data flow inside an oemolithread
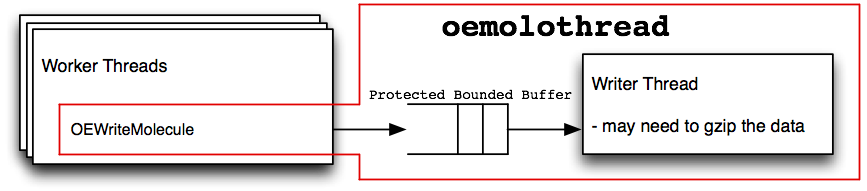
oemolothread
Diagram of data flow inside an oemolothread
Figure: oemolithread and Figure: oemolothread shows how data flows between an arbitrary number of worker threads and the I/O thread. Molecule data passes through a protected bounded buffer. It is “protected” against race conditions when multiple threads operate on the buffer. It is “bounded” in size to prevent increasing the required amount of memory by inserting many items into the buffer faster than can be consumed on the other side. The I/O threads will be put to sleep, allowing other threads to use the CPU, when there is not a sufficient amount of work for them to do. This means the number of worker threads should typically be the number of available CPUs. The I/O threads don’t require much CPU time, therefore, they don’t need their own dedicated CPU. This could change as the number of worker threads increases, or if the I/O thread is expected to handle data compression or uncompression.
Listing 2 demonstrates how to scale the
molecule searching program to
multiple CPUs. A worker thread is dedicated for each CPU. There are
also I/O threads to handle the input and output operations. The data
parsing (OEReadMolecule), substructure
searching(OESubSearch.SingleMatch), and data
serialization (OEWriteMolecule) is all done in
parallel by the worker threads. The costly synchronization point of
reading and writing data is handled by the separate I/O threads.
Listing 2: Using multiple threads to read and write to molthreads
package openeye.docexamples.oechem;
import java.util.ArrayList;
import openeye.oechem.*;
public class MultiCoreMolFind {
static OESubSearch ss;
static oemolithread ifs;
static oemolothread ofs;
public static class MolFindThread extends Thread {
public void run() {
OEGraphMol mol = new OEGraphMol();
while (oechem.OEReadMolecule(ifs, mol))
oechem.OEPrepareSearch(mol, ss);
if (ss.SingleMatch(mol))
oechem.OEWriteMolecule(ofs, mol);
}
}
public static void main(String argv[]) {
ss = new OESubSearch(argv[0]);
ifs = new oemolithread(argv[1]);
ofs = new oemolothread(argv[2]);
int ncpus = Runtime.getRuntime().availableProcessors();
ArrayList<MolFindThread> thrds = new ArrayList<MolFindThread>();
for (int i=0; i < ncpus; i++) {
MolFindThread thrd = new MolFindThread();
thrd.start();
thrds.add(thrd);
}
for (MolFindThread thrd : thrds) {
try {
thrd.join();
} catch (InterruptedException e) {
System.err.println(e);
}
}
ofs.close();
ifs.close();
}
}
Listing 2 will generate the same set of
molecules in the output file as Listing
1. However, there is big difference between the
output files, the molecule order is non-deterministic. For most users
this does not matter, database files are treated as arbitrarily
ordered sets of molecules. If the order of the molecule database is
important do not use the program in Listing
2.
Note
Database order is preserved by the program in Listing
1 because there is only one worker thread. This
allows users with database order restrictions to still take
advantage of threaded asynchronous I/O.
Thread Safety
OEChem TK is considered to be a “thread-safe” library. It makes the same guarantees the C++ Standard Template Library (STL) makes in concern of threads:
Unless otherwise specified, standard library classes may safely be
instantiated from multiple threads and standard library functions
are reentrant, but non-const use of objects of standard library
types is not safe if shared between threads. Use of non-constness
to determine thread-safety requirements ensures consistent
thread-safety specifications without having to add additional
wording to each and every standard library type.
Classes
Unless otherwise stated in the API documentation, OEChem objects can be instantiated and used in multiple
threads. For example, in Listing 2, special
care was taken to make the molecule object local to each thread.
This was accomplished by making an OEGraphMol
instance local to the thread’s run method. Therefore, ensuring
each thread had a separate OEGraphMol to read
molecules into.
public void run() {
OEGraphMol mol = new OEGraphMol();
while (oechem.OEReadMolecule(ifs, mol))
The same object can be accessed from multiple threads if all access is
through a const method. A const method is any method that does
not change data internal to the class. In C++, a method can be
explicitly marked const, allowing the compiler to cause the
code to not compile if this guarantee is not kept.
For example, the OESubSearch.SingleMatch method is
a const method. Therefore, the same OESubSearch
object can be used from multiple threads without the need to make
local copies of the OESubSearch object in
Listing 2. However, notice that the
molecule argument passed to OESubSearch.SingleMatch
is local to each thread. Otherwise, one thread could be reading data
into the molecule object while another thread is trying to perform the
substructure search, leading to a race condition and undefined
behavior.
if (ss.SingleMatch(mol))
Warning
Care must be taken when reusing the same objects in multiple
threads. Only one invocation of a non-const method
(e.g. OESubSearch.AddConstraint) concurrently
could crash the program. Concurrent execution of non-const
methods must to be protected by mutual
exclusion.
Functions
OEChem functions, unless otherwise
stated in the API documentation, can be safely called from multiple
threads. For example, OEGetTag needs to access a
global table to make string to unsigned integer associations
global to the program. OEGetTag ensures its own
thread safety so that it is safe to call from multiple threads.
However, mutual exclusion can be very expensive. This is why OEChem TK
(and the STL) make no effort to make concurrent object access
safe. So when mutual exclusion can not be avoided, as in the case of
OEGetTag, faster alternatives should be
sought. For example, the return of OEGetTag can
be cached to a local variable.
It is important to realize that even though the function may be
thread-safe, the function may be altering the state of its arguments,
which could be shared across threads. Again, the “const-ness” of the
arguments must be taken into account. For example, it is not safe to
call OEWriteMolecule on the same molecule from
separate threads (even on different
oemolostreams) because the molecule
argument to OEWriteMolecule is non-const. The
molecule argument is non-const for performance, the molecule may
need to be changed to conform to the file format being written
to. OEChem TK provides OEWriteConstMolecule that will
make a local copy of the molecule before making necessary changes for
the file format being requested.
Exceptions to the const argument rule must be explicitly stated in
the API documentation. For example, the
OEWriteMolecule function applied to the
non-const argument of oemolothread is thread-safe
only by virtue of design.
oechem.OEWriteMolecule(ofs, mol);
Memory Management
Memory allocation is often a bottleneck in any program. This is
especially the case in multi-threaded programs because memory
allocation must protect a globally shared resource, the memory space
of the process. Most malloc implementations use mutual exclusion
to make themselves thread-safe. However, mutual exclusion is
expensive, and cuts down on the scalability of the multi-threaded
program. OEChem TK attempts to alleviate the problem by implementing a
memory cache for the following small objects:
Any OpenEye Toolkit iterator object, i.e., something derived from OEIterBase.
Any OEGetNbrAtom functor. This is used by some implementations of
OEAtomBase.GetAtomsthat returns an iterator as well, see previous.The OEMatch class. Used by almost any API that operates with an instance of OEMatchBase.
OEMolBaseType.OEMiniMolinstances of OEGraphMol.
The cache is totally opaque to the normal OEChem TK user. However, it can cause problems in multi-threaded situations.
The small object cache implementation is controlled through the
OESetMemPoolMode function. The parameter passed to
this function determines what sort of cache implementation to use, the
possible options are a set of values bit-wise or’d from the
OEMemPoolMode constant namespace. The possible
combinations are as follows:
Warning
OESetMemPoolMode is not available in
Java. The reason is explained later.
- SingleThreaded|BoundedCache:
Makes no attempt at thread safety. Will only cache allocations up to a certain size, then further allocations will be sent to the global
operator new(size_t)function.- SingleThreaded|UnboundedCache:
Makes no attempt at thread safety. Will cache all allocations up to any size, never returning memory to the operating system after it has been requested.
- Mutexed|BoundedCache:
Protects a single global cache with a mutex. Will only cache allocations up to a certain size, then further allocations will be sent to the global
operator new(size_t)function.- Mutexed|UnboundedCache:
Protects a single global cache with a mutex. Will cache all allocations up to any size, never returning memory to the operating system after it has been requested.
- ThreadLocal|BoundedCache:
Uses a separate cache for each thread. Will only cache allocations up to a certain size, then further allocations will be sent to the global
operator new(size_t)function.- ThreadLocal|UnboundedCache:
Uses a separate cache for each thread. Will cache all allocations up to any size, never returning memory to the operating system after it has been requested.
- System:
Forwards all allocations to the global
operator new(size_t)function. This allows the user to circumvent the entire caching system in preference to their ownoperator new(size_t)function allowing other third party memory allocators to be linked against.
The default implementation in Java is Mutexed|UnboundedCache.
Unbounded for
performance, however,
Mutexed since the Java
garbage collector runs in a separate thread.
Thread Safety Options
Warning
The small object cache in Java OEChem TK must always be mutexed
since the garbage collector runs in another thread. This is why
OESetMemPoolMode is not exposed in the Java
interface.
If database order is a necessity it may be possible to gain
performance from threading using “pipeline”
parallelism. Listing 3 demonstrates how the
molecule searching problem can be broken down into 5 stages, each
running in a separate thread. In order, the 5 stages are:
Reading data from disk - oemolithread
Parsing the data into a molecule -
ParseThreadPerforming the substructure search on the molecule -
SearchThreadSerializing the molecule -
main threadWriting data to disk - oemolothread
Listing 3: Using pipeline parallelism in Java OEChem TK
package openeye.docexamples.oechem;
import java.util.concurrent.*;
import openeye.oechem.*;
public class PipelineMolFind {
static OESubSearch ss;
static oemolithread ifs;
static oemolothread ofs;
static BlockingQueue<OEGraphMol> iqueue;
static BlockingQueue<OEGraphMol> oqueue;
public static class ParseThread extends Thread {
public void run() {
try {
OEGraphMol mol = new OEGraphMol();
while (oechem.OEReadMolecule(ifs, mol)) {
iqueue.put(mol);
mol = new OEGraphMol();
}
mol.Clear();
// signal SearchThread to die with an empty molecule
iqueue.put(mol);
} catch (InterruptedException e) {
System.out.println(e);
}
}
}
public static class SearchThread extends Thread {
public void run() {
try {
OEGraphMol mol = iqueue.take();
while (mol.IsValid()) {
oechem.OEPrepareSearch(mol, ss);
if (ss.SingleMatch(mol))
oqueue.put(mol);
mol = iqueue.take();
}
mol = new OEGraphMol();
// signal main thread to die with an empty molecule
oqueue.put(mol);
} catch (InterruptedException e) {
System.out.println(e);
}
}
}
public static void main(String argv[]) {
ss = new OESubSearch(argv[0]);
ifs = new oemolithread(argv[1]);
ofs = new oemolothread(argv[2]);
iqueue = new ArrayBlockingQueue<OEGraphMol>(1024);
oqueue = new ArrayBlockingQueue<OEGraphMol>(1024);
Thread pthrd = new ParseThread();
pthrd.start();
Thread sthrd = new SearchThread();
sthrd.start();
try {
OEGraphMol mol = oqueue.take();
while (mol.IsValid()) {
oechem.OEWriteMolecule(ofs, mol);
mol = oqueue.take();
}
} catch (InterruptedException e) {
System.out.println(e);
}
ofs.close();
ifs.close();
}
}
Overriding the allocator
Memory pooling is even more important in Java because all OpenEye Toolkit memory allocation actually allocates memory from the JVM heap. Java maintains strict control over the amount of heap memory the JVM should use through the -Xmx flag. In order for the OpenEye Toolkits to adhere to these limits and put pressure on the garbage collector, all OpenEye Toolkit memory is allocated through the JNI as byte array objects. This makes small object memory caching even more important for Java as there is a higher overhead in both space and time for each memory allocation.
OpenEye testing has shown that the current default of
Mutexed|UnboundedCache to be a good balance of performance and
memory usage. However, it is not particularly suited to heavily
multi-threaded Java programs due to the use of a mutex. While
OESetMemPoolMode is not available in Java
because it is very race condition prone inside the JVM, there are
the following environment variables that can be set to experiment
with a better memory strategy:
OEJAVA_BOUNDED_CACHEOEJAVA_MEM_POOL_MODE
OEJAVA_BOUNDED_CACHE specifies that the memory pool mode should
be Mutexed|UnboundedCache to be more aggressive about returning
memory to the operating system.
OEJAVA_MEM_POOL_MODE can take any integral value that
OESetMemPoolMode will accept. The user will
have to go investigate the oesystem/threadsafe.h to figure out
the right integer. The most common one is to use the integer 16
for OEMemPoolMode.System to bypass all small
object caching.
Warning
The use of these variable is intentionally difficult. This is an area of active research at OpenEye and we hope to remove them entirely in the future with a better default mode that works well for both multi-threaded and single threaded Java programs. Encouraging results have been seen by tighter integration with TCMalloc, and these variable are only here to encourage such experimentation.
(Un)bounded Options
Two types of small object caches can be set with
OESetMemPoolMode:
Bounded and
Unbounded. The default
is an Unbounded cache
for performance. The assumption is most OEChem TK programs will have a
constant memory usage. Even if the program has a steadily rising
memory footprint the program will usually exit after it has maxed out
its memory requirements.
This may not be the case for long running programs, for example, a web
service. These programs may need to create a temporarily large memory
footprint and then release it back to the system for some other
program to use. An
Unbounded cache
will hold onto all memory it sees, never returning any to the
operating system.
A Bounded cache will
only cache allocations up to a limit. After that limit has been
reached all further allocations will be sent to the system memory
allocator. This allows the deallocations later in the program to
return the memory to the operating system. This allows certain objects
to still use the fast cache while the other objects that are bloating
the memory footprint will incur the cost of asking the operating
system for the memory they require.
Note
Since OESetMemPoolMode is not available in
Java the bounded cache is selected by turning on the
OEJAVA_BOUNDED_CACHE environment variable.