OEGet2dPSA
OEGet2dPSA(OEMolBase mol, OEFloatArray atomPSA=None, bool SandP=False) -> float
Returns the topological polar surface area for a given molecule as
described in the Polar Surface Area section. The
SandP parameter controls whether sulfur and phosphorus should be
counted towards the total surface area.
See example in Figure: Example of depicting the atom contributions of
the polar surface area.
Warning
TPSA values are mildly sensitive to the protonation state of a molecule.
The atomPSA parameter can be used to retrieve the contribution
of each atom to the total polar surface area as shown in
Listing 1.
Listing 1: Example of retrieving individual atom contributions to PSA.
atomPSA = oechem.OEFloatArray(mol.GetMaxAtomIdx())
psa = oemolprop.OEGet2dPSA(mol, atomPSA)
print("PSA =", psa)
for atom in mol.GetAtoms():
idx = atom.GetIdx()
print(idx, atomPSA[idx])
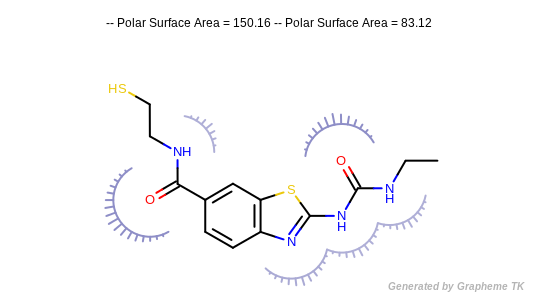
Example of depicting the atom contributions of the polar surface area (ignoring S and P atoms)
(Darker colors and longer spikes indicate larger PSA atom contributions)
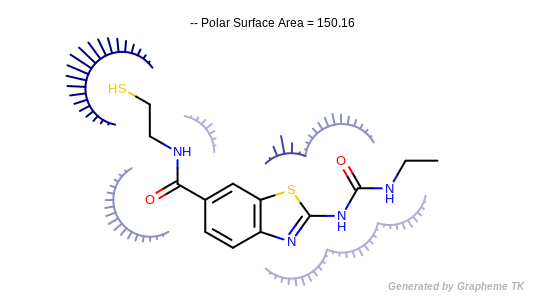
Example of depicting the atom contributions of the polar surface area (considering S and P atoms)
(Darker colors and longer spikes indicate larger PSA atom contributions)
See also
The Python script that visualizes the polar surface area of a molecule can be downloaded from the OpenEye Python Cookbook
See also
Polar Surface Area section