Generate database for fast substructure search
A program that generates a binary file for fast substructure search.
For more explanation, see OECreateSubSearchDatabaseFile function.
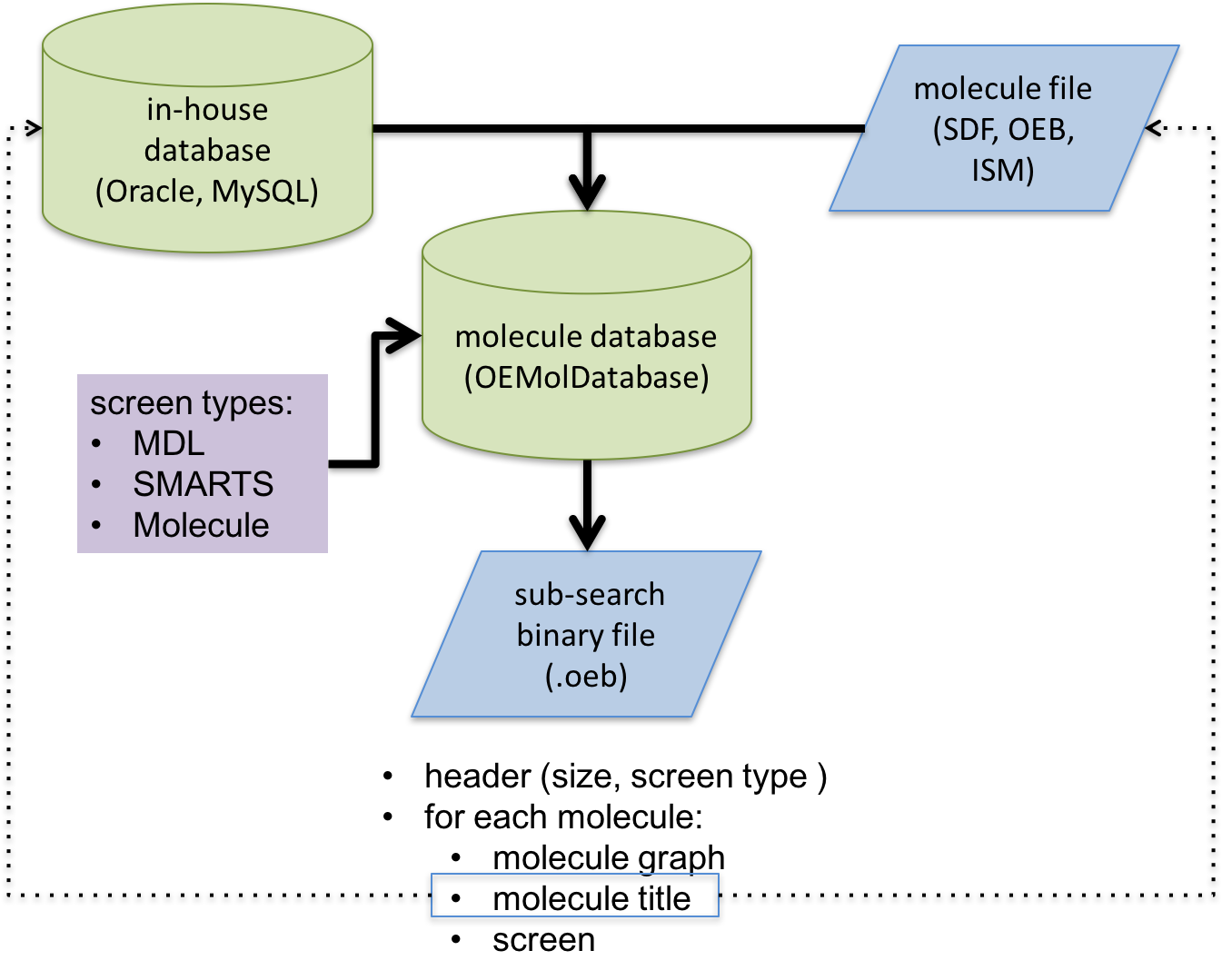
Schematic representation of substructure search database file generation process
Code Example
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> makefastss --help
will generate the following output:
Simple parameter list
input/output options
-in : Input molecule filename
-out : Output substructure database filename
other options
-keep-title : Whether to keep the title of the molecule as unique
identifier
-nrthreads : Number of processors used (zero means all available)
-screentype : Screen type generated
-sort : Whether to sort the molecules based on their screen bit counts.
Example
prompt> makefastss -in dataset.ism -out ss-search-dataset-SMARTS.oeb -stype SMARTS
Code
Download code
See also
OEConsoleProgressTracer class
OEGetSubSearchScreenTypefunctionOECreateSubSearchDatabaseFilefunctionOECreateSubSearchDatabaseOptionsfunction