OECreateSubSearchDatabaseFile
Attention
This API is currently available in C++ and Python.
bool OECreateSubSearchDatabaseFile(const std::string &ofname,
const std::string &ifname,
const OESubSearchScreenTypeBase *stype,
OESystem::OETracerBase &tracer=OESystem::OENoTracer)
bool OECreateSubSearchDatabaseFile(const std::string &ofname,
const std::string &ifname,
const OECreateSubSearchDatabaseOptions &opts,
OESystem::OETracerBase &tracer=OESystem::OENoTracer)
Generates substructure search database files for OESubSearchDatabase.
OECreateSubSearchDatabaseFile and returns true if the database
is successfully created.
- ofname
The name of the generated substructure search database file. The generated file will be an OpenEye binary file (
OEFormat::OEB); therefore,.oebfile extension is required in the given filename.- ifname
The name of the input molecule file.
- stype
The type of the screen that will be pre-generated to each molecule.
- opts
The OECreateSubSearchDatabaseOptions object that encapsulates properties that determine how the database is generated.
- tracer
A tracer that can be used to report the progress of the database generation.
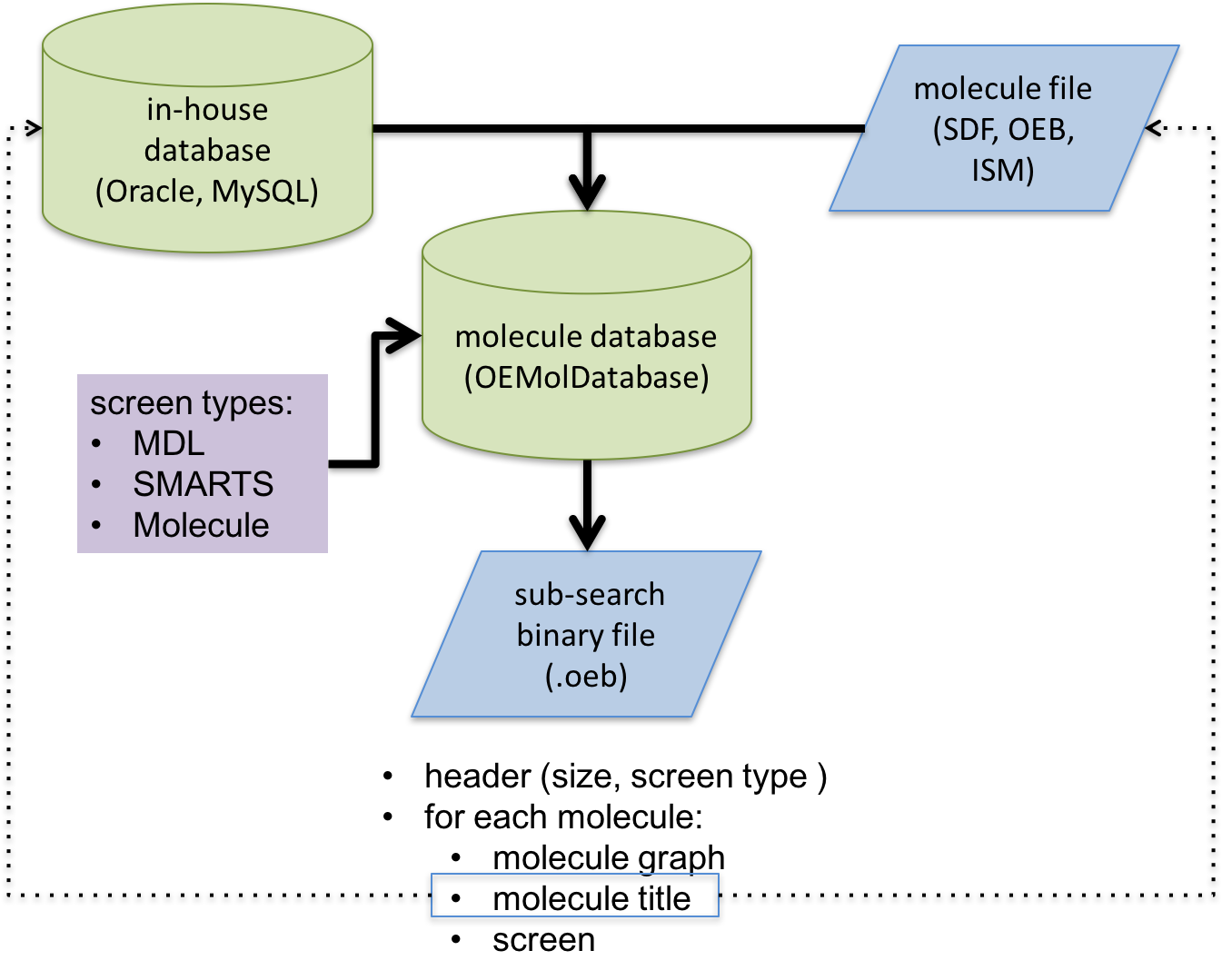
Schematic representation of substructure search database file generation process
The generated database file is an OpenEye binary file with a header that stores extra information about the database such as, screen type. Each substructure database file is associated with one and only one screen type. The molecules will be pre-processed for that specific type of substructure search. In order to minimize the memory footprints of the dataset at search time, all unessential data will be stripped from the molecules and explicit hydrogens will be suppressed. The search database will store only the following information for each molecule:
the molecular graph (no coordinates)
the molecule title (either user-defined or a unique identifier assigned during database generation)
the screen generated for the molecule
The titles are kept in order to provide an external id for each molecule.
See also OECreateSubSearchDatabaseOptions::SetKeepTitle.
When searching the database, the titles of the matched molecules can be
retrieved by the OESubSearchDatabase::GetMatchTitles method.
Note
If a sorted database is generated (see the
OECreateSubSearchDatabaseOptions::SetSortByBitCounts method),
a temporary file will be written into the disk prior to generating the final
sorted database. This two-step process allows to generate database files that
can not be fit into the memory with the size that is limited by the disk
space.
See also
OEConsoleProgressTracer class
Code Example