Perform SMARTS search on substructure database file
A program that searches substructure search database using a SMARTS pattern.
For more explanation, see OESubSearchDatabase class.
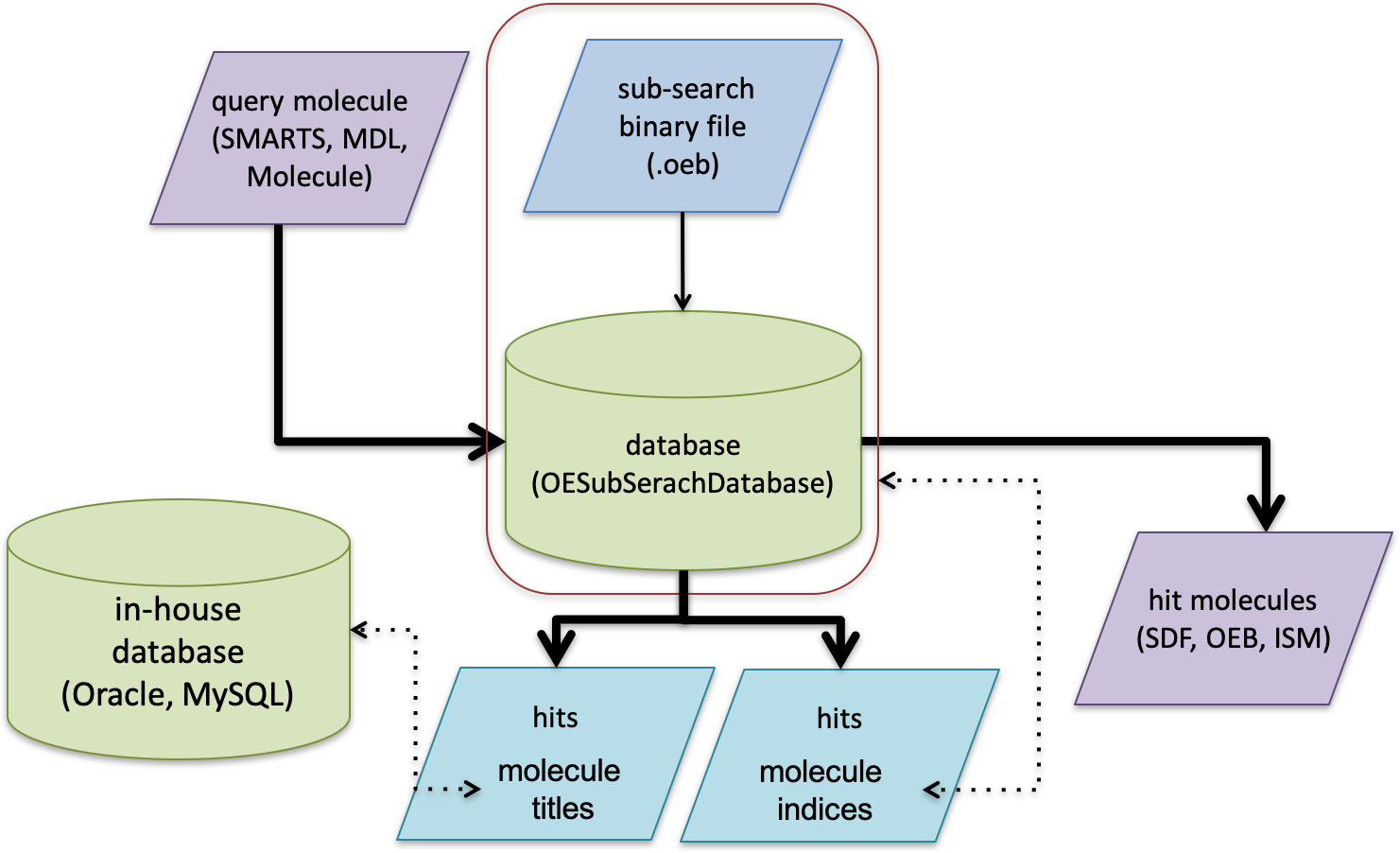
Schematic representation of using substructure search database
Code Example
Command Line Interface
A description of the command line interface can be obtained by executing the program with the –help argument.
prompt> searchfastss --help
will generate the following output:
Simple parameter list
input/output options
-database : Input substructure search database filename
-smarts : SMARTS pattern, quote for safety
other options
-maxmatches : The maximum number of matches returned in case of parameters
(-titles) and (-out)
-nrthreads : Number of processors used (zero means all available)
search types
-count : Output count of number matched
-out : Output molecule filename of matches
-titles : Output title of matches
Example
prompt> searchfastss -db ss-search-dataset-SMARTS.oeb -s "C1CC[O,N]CC1" -out matches.ism
prompt> searchfastss -db ss-search-dataset-SMARTS.oeb -s "C1CC[O,N]CC1" -count
prompt> searchfastss -db ss-search-dataset-SMARTS.oeb -s "C1CC[O,N]CC1" -titles
Code
Download code
See also
OESubSearchDatabase class
OESubSearchQuery class
OESubSearchResult class
OEConsoleProgressTracer class
OEGetSubSearchScreenTypefunctionOEIsValidSubSearchDatabasefunctionOESubSearchStatusToNamefunction