OEToolkits 2016.Feb
OpenEye Toolkit Security
OpenEye toolkits are increasingly being used in web services that require protection from malicious users. The most obvious attack vector against the OpenEye toolkits is file format parsing since scientific file formats are complex and often underdefined.
OpenEye has closed a significant number of vulnerabilities related to the parsing of supported file formats: molecules, grids, surfaces, receptors, etc. In addition, state-of-the-art testing and detection techniques are now continuously run on the OpenEye code base to ensure a high level of security going forward.
Security is a task requiring continuous vigilance and OpenEye will continue to make this effort a high priority.
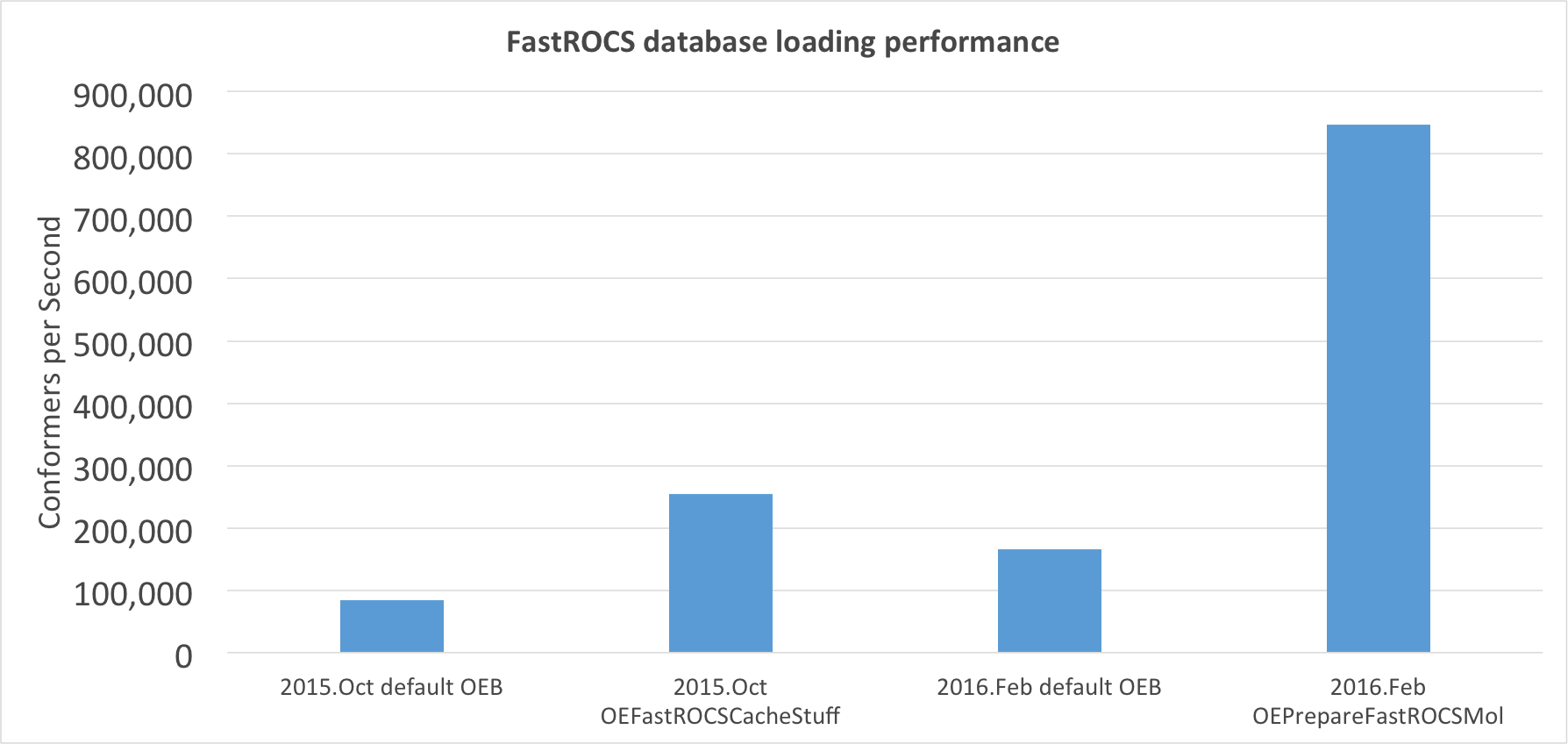
FastROCS TK: Database Loading Performance Improvement
The utility of FastROCS has always been hampered by the amount of physical memory available in GPU machines. The computational chemistry community is continuously pushing the boundaries of FastROCS by searching larger and larger virtual libraries of compounds. In addition, FastROCS servers are becoming an increasingly important component of many infrastructures and must be able to be rapidly brought up and down on demand. Both these problems come down to the fact that loading a database into memory is often between 10 and 20 times slower than performing an actual FastROCS search on that database.
This problem has been addressed in this release by dramatically increasing the
performance when loading a dataset into memory for eventual FastROCS processing.
OEPrepareFastROCSMol generates additional information to be written
to OEB that is used to rapidly read molecules into memory.
Using OEPrepareFastROCSMol on a molecule can result in
load times that are nearly 10 times faster than previous uncached OEB file loads.
Depending on the ratio of CPU and GPU compute power and conformers per molecule,
this can mean that loading a dataset can now be just as fast as the FastROCS
search itself.
|
OEMedChem TK
OpenEye is pleased to announce the first official release of OEMedChem TK. This medicinal chemistry functionality was previously only available in beta form.
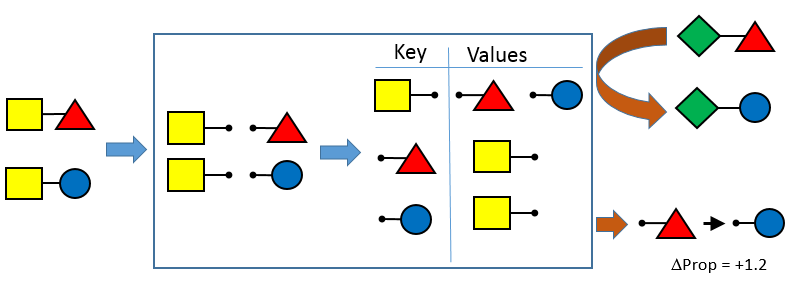
OEMedChem TK provides the ability to index a set of input structures and identify matched molecular pairs. Matched molecular pair analysis is becoming recognized as a powerful tool for the extraction of the effects of chemical changes on a property or properties of interest in large data sets.
|
Additional OEMedChem TK capabilities include fragmentation and perception, similarity measures, belief theory, and molecular complexity measures. For the full list of capabilities, see the OEMedChem functionalities section.
Note
If you would like to utilize the functionalities of OEMedChem TK, please visit https://www.eyesopen.com/contact for a license.
General Notices
This 2016.Feb release is the last to support Ubuntu 12 for all toolkits.
OSX 10.8 is no longer supported, but support has been added for OSX 10.11.
This 2016.Feb release is the last to support Visual Studio 2012 for C++ and C# toolkits and Visual Studio 2010 for Python 3.3 toolkit.
This 2016.Feb release supports Python 3.5 for the following platforms: OSX 10.10, OSX 10.11, Ubuntu 12, Ubuntu 14, RedHat 6, and RedHat 7.
The 2016.Jun release is expected to support Python 3.5 on Windows with Visual Studio 2015.
The 2016.Jun release will add C++ support for Visual Studio 2015.
The Python 2 distributions of OpenEye Python Toolkits can now handle
both ascii and unicode Python strings for all APIs that accept string arguments.
For example, the OESmilesToMol function can now parse
the following SMILES strings without throwing a ‘TypeError’ exception:
smiles_str = "[N+](=O)([O-])([O-]) nitrát"
smiles_unicode = u"[N+](=O)([O-])([O-]) nitrát"
Previously, the unicode strings would work in Python 3, but not in Python 2, making it difficult to write code that was compatible with both major versions of Python at the same time. This major change should make porting to Python 3 much easier for OpenEye toolkit users.